Highlights of our Work
2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
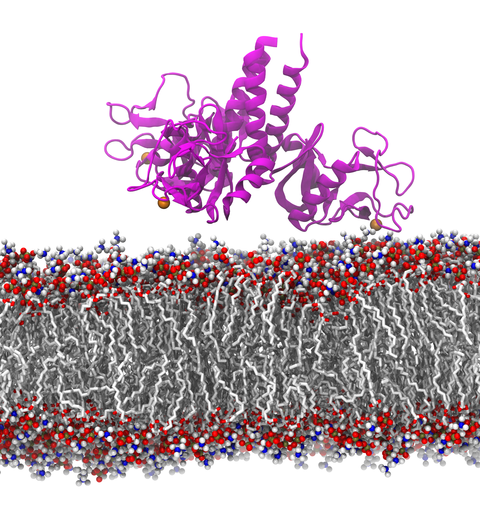
image size:
1.7MB
made with VMD
Our lungs are coated with a layer of protein and lipid mixture called lung surfactant, which prevents the lungs from collapsing and protects us from bacterial and viral infections
(see October 2012 and January 2014 highlights).
Lung surfactant protein A (SP-A) - the major protein constituent of lung surfactant -
plays a dual role.
It aggregates DPPC lipid, a major component of lung membrane,
into a lattice-like structure that prevents the lungs from collapsing.
SP-A is also known to recognize and bind bacterial lipids, namely lipid A, on surfaces of gram-negative bacteria,
thereby helping to initiate various clearance mechanisms.
However, it was unclear how SP-A exhibits such functional duality with its binding to two different types of lipids.
A recent study
used molecular
dynamics simulations with
NAMD to unravel the dual role of SP-A.
Combined with crystallographic and mutational analyses,
researchers have discovered several critical, non-canonical lipid binding sites that
involves cation-π interactions and hydrogen bonds.
Simulations have also revealed that SP-A binds stronger to bacterial lipid (lipid A)
than to surfactant lipid (DPPC lipid), which suggests SP-A may prioritize its host defense
functions by transferring from lung membrane to bacterial surface.
These findings in atomistic detail will enable experimentalists to enhance the antimicrobial function of SP-A.
More on our lung surfactant protein website.



