Driving Biomedical Projects
A Driving Biomedical Project (DBP) involves research in our group that poses a major technical challenge and requires significant further development of our existing software tools, or even entirely new software tools. Below are descriptions of DBPs currently pursued in our group.
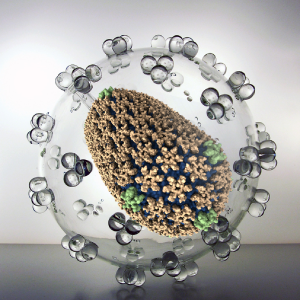
Viruses represent a serious public health threat and millions of people die every year from viral diseases. In an effort to prevent and combat viral infection, researchers worldwide are endeavoring to develop vaccines and drug-based treatments, particularly targeting virus capsids. All-atom description of virus capsids has proven to be extremely valuable in accurately characterizing protein-protein interfaces and in revealing novel interactions between capsids and host factors / drug molecules. Here, innovations in computational modeling and simulation techniques will enable the study of (1) the effect of drug molecules on virus capsids and (2) the dynamic structure of RNA inside capsids.
Read more about the Center's virus research and the five year research plan for DBP1.
Collaborating Investigators: A. Gronenborn (U. Pittsburgh); P. Zhang (U. Pittsburgh); C.R. Aiken (Vanderbilt U.); T. Polenova (U. Delaware); S.G. Sarafianos (U. Missouri); A. Zlotnick (Indiana U.); R.C. Craven (Penn. St. U.); I. Hofacker (U. Vienna)
Funding: NIH P50-GM082251: 08/2007-06/2017 (Polenova, Aiken, Zhang and Gronenborn); NIH R01-AI067417: 12/2005-05/2017 (Zlotnick); NIH R01-AI118933: 03/2016-02/2021 (Zlotnick); NIH R01-AI120860: 06/2015-05/2020 (Sarafianos); NIH R01-AI071286: 04/2012-03/2017 (Craven); DACH project Austrian Science Fund: 04/2014-03/2017 (Hofacker)
Key Publications: Perilla et al., Journal of Physical Chemistry Letters, 2016; Liu et al., Nature Communications, 2016; Zhao et al., Nature, 2013
Symbiont bacteria greatly influence human health and play a significant role in pathogenesis, disease predisposition, physical fitness, and dietary responsiveness. Here we propose to investigate two key processes underlying bacterial interactions with humans: chemotaxis and plant fiber metabolism. Specifically, we will study the structure and function of the highly cooperative macromolecular complexes central to each of these processes, namely the chemosensory array, a transmembrane cluster of sensory proteins, and the cellulosome, a large extracelular enzymatic complex. Innovations in computer simulation techniques will be used to investigate, in particular, the molecular origins of (1) robust sensory signal transduction within the chemosensory array and (2) highly efficient plant fiber degradation by cellulosomes.
Read more about the Center's symbiont bacteria research and the five year research plan for DBP2.
Collaborating Investigators: I. Cann (UIUC); E. Bayer (Weizmann); H. Gaub (LMU Munich); P. Zhang (U. Pittsburgh); S. Parkinson (U. Utah); M. Eisenbach (Weizmann); A. Briegel (Leiden U.); M. Nash (U. Basel); J. Ridlon (UIUC);
Funding: NSF/PFC/CPLC PHY1430124: 09/2008-07/2019 (Schulten); EBI: 2008-2016 (Cann); ISF 1349/13: 2013-2018 (Bayer); ERC 294438: 03/2012-02/2018 (Gaub); NIH R01-GM085043: 08/2009-07/2016 (Zhang); NIH R01-GM1955943: 05/2015-02/2019 (Parkinson); ISF 534/10: 2014-2018 (Eisenbach); Leiden U. startup funds (Briegel); HFSP Young Inv.: 12/2015-05/2019 (Nash); Branco Weiss: 05/2013-05/2018 (Nash)
Key Publications: Cassidy et al., eLife, 2015; Schoeler et al., Nature Communications, 2014; Schoeler et al., Nano Letters, 2015
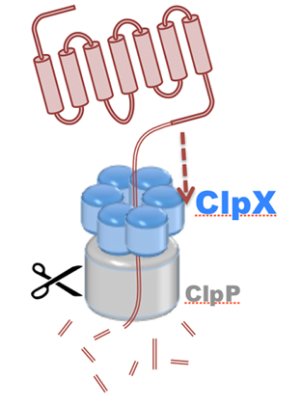
Motor proteins are molecular machines that convert chemical energy from ATP hydrolysis into mechanical work to drive conformational transitions regulating a range of biological processes. Defects in motor protein function are implicated broadly in cancer, as well as numerous cardiovascular, neurological, and reproductive diseases. The chemo-mechanical properties of motor proteins remain poorly characterized for the majority of systems, primarily due to the structural complexity of molecular machines, as well as the long (millisecond) timescales of their action. Here, innovations in computer simulation techniques will enable investigation of the mechanisms of hydrolysis-driven motion underlying (1) regulation of ATP synthesis by ATP synthase, (2) protein degradation by ClpX unfoldase, and (3) cytoskeletal cargo transport by cytoplasmic dynein.
Read more about the Center's molecular motors research and the five year research plan for DBP3.
Collaborating Investigators: Y. Goldman (U. Penn); A. Martin (U.C. Berkley); J. Rubinstein (U. Toronto)
Funding: NSF/PV&V MCB-1157615 (Schulten); NIH 5P01GM087253-12: 08/2015-07/2016 (Goldman); CIHR MOP-81294: 10/2014-09/2019 (Rubinstein); NSERC Discovery Grant: 04/2012-03/2017 (Rubinstein); NIH R01 GM 09447-05: 07/2011-06/2016 (Martin); NSF-MCB-1150288: 09/2012-08/2017 (Martin)
Key Publications: Ma et al., JACS, 2015; Moradi et al., Nature Communications, 2015
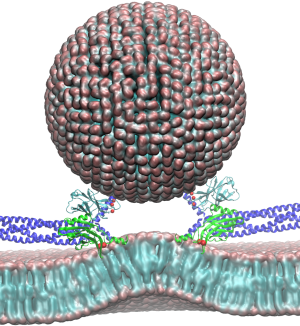
The brain is the source of thoughts, perceptions, emotions, memories and actions. Neural signaling, the foundation of brain activity, must be precisely regulated to prevent neuronal disorders that may cause Parkinson's disease, schizophrenia, compulsive behaviors and addiction. Such a precise regulation is achieved by key signaling proteins, voltage-gated sodium (Na+) and potassium (K+) channels for electrical signaling and calcium (Ca2+) - bound synaptotagmin for chemical signaling. Here, innovations in computer simulation techniques will be used to investigate the molecular mechanism of neural firing induced by voltage-gated Na+ and K+ channels and membrane fusion triggered by synaptotagmin.
Read more about the Center's neurons and synapses research and the five year research plan for DBP4.
Collaborating Investigators: C. A. Ahern (U. Iowa); Y. Shin (Iowa State U.); A. Brunger, (Stanford U.)
Funding: NSF/PFC/CPLC PHY1430124: 09/2008-07/2019 (Schulten); NIH R01-GM106569: 07/2013-06/2018 (Ahearn); NIH U54-GM087519: 06/2015-05/2020 (Schulten, Ahern, Shin); NIH R01-GM051290: 08/2015-04/2020 (Shin); NIH R37-MH063105: 03/2016-02/2021 (Brunger)
Key Publications: Wu et al., Biophysical Journal, 2014; Blanchard et al., Biophysical Journal, 2014; Khalili-Araghi et al., Biophysical Journal, 2010
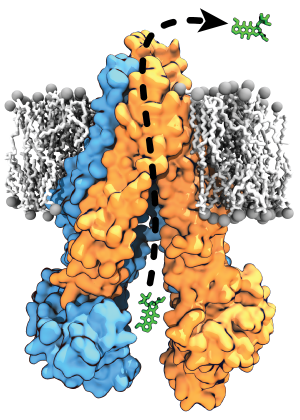
Membrane transporters perform the fundamental biological task of efficient and selective transport of materials across the cell membrane using diverse forms of cellular energy. Many transporters have been identified as drug targets, and effective drug development necessitates a comprehensive understanding of their structural intermediates and transitions during the transport cycle. The Center has developed a set of advanced methodologies combining extensive sampling of the phase space and free energy calculations to structurally and thermodynamically characterize the complete transport cycles of the transporters. Here, further development of this methodology will enable structural characterization of transport cycle of two structurally and mechanistically diverse classes of membrane transporters: P-glycoprotein (P-gp), a primary (ATP-driven) transporter implicated in multidrug resistance in cancer, and VcINDY, a secondary (ion-driven) transporter mediating transport of metabolic intermediates.
Read more about the Center's five year research plan for DBP5.
Collaborating Investigators: H. Mchaourab (Vanderbilt); R. Nakamoto (U. Virginia); D.-N. Wang (NYU)
Funding: NIH U54-GM087519: 08/2010-08/2018 (Mchaourab, Nakamoto, Tajkhorshid); NIH R01-DK099023: 03/2013-02/2017 (Wang); NIH R01-GM086749: 08/2009-07/2016 (Tajkhorshid)
Key Publications: Wen et al., Journal of Biological Chemistry, 2013; Mishra et al., eLife, 2014
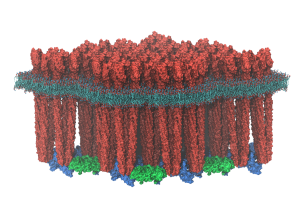
Respiration and photosynthesis are ubiquitous bioenergetic processes for harvesting and converting energy in biological cells. Many key proteins and physical processes are common to both processes, featuring organelle-scale arrays of membrane protein complexes. Dysfunction of the repiratory complex is associated with several pathologies, including Parkinson's disease, malaria, and breast cancer. Here, molecular modeling techniques from single-protein to organelle scales are employed to study (1) energy conversion in a respiratory supercomplex, called respirasome, in mitochondria and (2) energy harvesting by a granal thylakoid membrane in chloroplasts.
Read more about the Center's bioenergetic membranes research and the five year research plan for DBP6.
Collaborating Investigators: N. Hunter (U. Sheffield); R. Gennis (U. Illinois); L. Sazanov (IST Austria ); R. Blankenship (Wash. U.)
Funding: NSF/PFC/CPLC PHY1430124: 09/2008-07/2019 (Schulten); NSF MCB-1157615: 5/1/12-4/30/16 (renewal submitted) (Schulten); DOE DE-SC0001035: 8/1/14-7/31/18 (Blankenship, Hunter, Schulten); NSF NSF-1616874: 07/15/16-06/15/20 (Gennis); IST start-up funds: 02/01/2014-01/01/2018 (Sazanov)
Key Publications: Stone et al., Parallel Computing, 2016; Singharoy et al., JACS, 2016; Sener et al., eLife, 2016
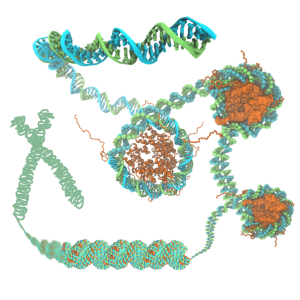
The structure of chromatin plays a pivotal role in gene expression in eukaryotic cells, but nevertheless remains poorly characterized. The basic unit of chromatin is a nucleosome particle formed by wrapping double-stranded DNA around histone proteins. Nucleosomes interact with each other in part through disordered tails. Histone tails are targets for epigenetic modifications that alter chromatin compaction and, correspondingly, gene expression. The structure of chromatin is of particular interest since chromatin remodeling genes, which affect chromatin structure, are implicated in a number of diseases (e.g. Rett syndrome, Rubinstein-Taybi syndrome, alpha thalassemia) and in the genesis of many types of cancer. Innovations in computer simulation techniques that combine atomic and coarse-grained (CG) methods will be used to (1) elucidate how physical forces drive chromatin organization and (2) determine the effects of epigenetic modifications on chromatin structure.
Read more about the Center's five year research plan for DBP7.
Collaborating Investigators: T. Ha (JHU); J. Song (UIUC)
Funding: NSF/PFC/CPLC PHY1430124: 09/2008-07/2019 (Aksimentiev, Ha, Song); HHMI: 9/2005--8/2016 (Ha); NIH R01 GM112659-01: 1/2015--11/2018 (Ha); Sontag Foundation: 10/2011--8/2017 (Song); NIH R01 CA163336-01: 12/2011--12/2016 (Song)
Key Publications: Yoo et al., Nature Communications, 2016; Yoo et al., Nucleic Acids Research, 2016; Maffeo et al., Journal of Chemical Theory and Computation, 2014
Hematopoietic stem cells (HSC) are responsible for maintaining the body's blood and immune cells; dysregulation of their behavior can lead to severe pathologies like leukemia. Understanding their proliferation and diffentiation could not only shed light on pathological processes, but could also help to expand HSC populations for transplantations. However, such understanding requires the study of gene expression and regulation, of signaling within the highly compartmentalized eukaryotic intracellular space, and of cellular interactions with complex environments - processes that can be investigated in simpler model systems. Here, we will study (1) LeuRS:tRNA-mediated intercellular signaling in Escherichia coli as a model for signaling by cytokines, especially stem cell factor (SCF), (2) the spliceosome assembly in Saccharomyces cerevisiae as a model for multi-compartmental cellular processes, and (3) HSC proliferation and differentiation in an in vitro niche environment.
Read more about our Center's five year research plan for DBP8.
Collaborating Investigators: W. Baumeister (MPI Munich); E. Villa (UCSD); M. Gruebele (UIUC); S. Martinis (UIUC); B. Harley (UIUC) H. Nelson (Mayo)
Funding: WM Keck Foundation 206231:01/2015-11/2019 (Martinis); NSF MCB 12-44570 (Luthey-Schulten); NSF 125738 (Harley); NIH R01DK099528: 08/2014-07/2019 (Harley); Am. Cancer Soc. 189782 (Harley); NIH R01 GM093318-04-08: 09/2013-8/2017 (Gruebele); U. California (Villa); MPG (Baumeister)
Pending Funding: NIH R01 (Nelson/Luthey-Schulten)
Key Publications: Earnest et al., Biopolymers, 2016; Cole et al., BMC Systems Biology, 2015; Earnest et al., Biophysical Journal, 2015
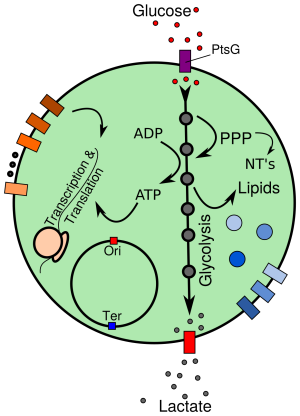
All cells share a universal, minimal set of biochemical processes, essential to all life. The search for defining this minimal set has lead to the synthetic assembly of the first minimal cell, JCVI-syn3.0: a cell with only 473 genes, the bare minimum requirements for independent cellular life. Its genetic basis was Mycoplasma mycoides; the Mycoplasmas have long been of interest to investigate fundamentals of life due to their evolutionarily reduced genomes, and at the same time represent an important class of pathogens, implicated in diseases such as pneumonia, urogenital diseases and certain types of cancer. Thus, JCVI-syn3.0 constitutes a platform to study the function of every gene that is essential for cellular life, and provides simultaneously a prototype for investigating cellular networks in a number of pathogens. The small (sub-micrometer) size and simple genome of JCVI-syn3.0, together with the timely availability of proteomics and cryo-electron tomography (cryo-ET) data in progress, bring an in silico model of the entire minimal cell at all-atom resolution within reach. This model will enable a spatially resolved study of the biochemical processes fundamental to life. At the same time, it will allow the computational analysis of cell-scale effects resulting from biomedically relevant perturbations (e.g., antibiotics) on this pathogen prototype. Here, using the proposed technological advances of the Center, as well as planned exascale machines, we will study: (1) the complete, minimal network of cellular processes, i.e. DNA replication, transcription, translation, ribosome assembly and metabolism, (2) the physical properties of the crowded cytoplasm and cell membrane within the entire JCVI-syn3.0 cell, and their coupling to the network of cellular processes and (3) biomedical applications, including: molecular and cell-scale effects of antibiotics; the evolution of new metabolic functions, to reconstruct the emergence of virulence; and the potential implementation of new biosynthetic pathways to produce biomedical compounds of interest.
Read more about the Center's five year research plan for DBP9.
Collaborating Investigators: C. Hutchison (JCVI); E. Villa (UCSD)
Funding: DARPA HR0011-12-C-0063: 05/17/2012-11/10/2014 (Hutchison); SGI (Hutchison); JCVI (Hutchison); U. California (Villa)
Pending Funding: NIH R01 (Hutchison/Villa/Luthey-Schulten), MacArthur (Hutchison/Villa/Luthey-Schulten)
Key Publications: Goh et al., Annual Review of Biophysics, 2015; Perilla et al., Current Opinion in Structural Biology, 2015