Molecular Modeling in the Cloud
The use of advanced molecular simulation techniques often comes with additional computational and software requirements. Running molecular simulation and analysis tasks in the Cloud can significantly lower the barriers to use of advanced simulation methods and provides a cost-effective and practical solution for many molecular modeling tasks and for small and moderate size molecular dynamics simulations.
Cloud computing involves the use of computational equipment provided by a third-party, accessed through an internet connection, and paid for on an on-demand, as-needed basis. The cloud computing model provides researchers with access to powerful computational equipment that would otherwise be too costly to procure and maintain on their own. Tasks such as MDFF structure refinement often involve the use of multiple software suites, such as VMD for simulation preparation and Situs for initial rigid-body docking and VMD, NAMD, and ROSETTA for iterative refinement of models. Cloud platforms are also useful for bundling together all of the software packages used in a modeling workflow to gauruntee their availablity and interoperability on a standardized system.
We have adapted our molecular modeling applications NAMD, VMD,
and associated tools to operate within the Amazon Web Servers (AWS)
Elastic Compute Cloud (EC2) platform, enabling popular research workflows
such as MDFF structure refinment and QwikMD simulation protocols to be
run remotely by scientists all over the globe, with no need for
investment in local computing resources and a reduced requirement for
expertise in high performance computing technologies.
High Performance Hardware At Your Fingertips
Maximize Your Efficiency Running VMD and NAMDAWS EC2 has a wide variety of 'instance types' backed by some of the latest high performance CPU and GPU hardware. These instance types give you access to a level of performance that can not be found in your typical personal desktop or laptop. This is perfect for the researcher with limited access to, or need for, expensive computational equipment and removes the need for maintaining hardware. You'll never have to worry about replacing a broken hard drive. Hardware details for each EC2 instance type can be found here, and instance pricing is listed here. |

|
Recommended Instance TypesVisualization and GPU Acceleration For visualization work, we recommend running VMD on GPU-accelerated EC2 instance types such as g3s.xlarge, p3.2xlarge. The "g3" GPU-accelerated instances use Tesla M60 GPUs that perform well for general VMD visualization and analysis tasks. The "p3" GPU-accelerated instances use Tesla V100 GPUs, and are much more powerful and are better suited for large scale rendering and analytical workloads in VMD or simulation of smaller structures with NAMD, due to their comparatively large 16GB on-board GPU RAM. General Computation For some simulations or analyses which are not GPU accelerated or are performance bound by the CPU, access to many CPU cores on a single node, or across a cluster of nodes (advanced usage), may be preferred. In these cases we recommend the compute optimized C4 or C5 instance types, which can give you up to 72 CPUs on a single node.
|
Trajectory or Structure Analysis For work largely dominated by I/O, we recommend using instance types that support high performance networking such as "g3", "p3" (GPU-accelerated) or "m4", "c5", "r4" (CPU-only). In some cases, the use of instance-local SSDs for temporary storage may be beneficial for analysis tasks, and for those cases we would suggest using the "C5d" instances. |
Remote Visualization
Access Your Data From AnywhereWith the power of NICE DCV, you can seamlessly connect to your running EC2 session and smoothly interact with it as if you were sitting at the machine itself. You can harness the powerful EC2 hardware from your personal desktop or laptop, and run demanding simulations or analyses from anywhere with an internet connection |
|
|
Advance Visualization and Movie and Image RenderingUsing the g3 instance types and DCV, you can use the most advanced visualization features of VMD, like interactive ray tracing to render stunning images and movies. |
Easily Access the Cloud
Launch With a Single ClickOur cloud virtual machine image is availble in the Amazon Marketplace, which allows users to use a very simple one-click launch using pre-configured choices of instance types that have been tested with our software. No further special configuration or setup is necessary |
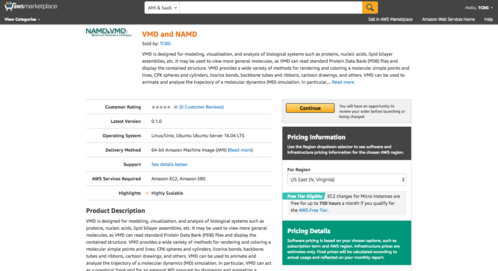
|
|
Remote Visualization with DCV If you are running on a g3 instance type, you can graphically access your instance with NICE DCV. To access a running instance, you will need to use its public IP. The password is the instance id of the running instance. Once connected to the instance, you may run NAMD or VMD from the command line as usual, or run VMD from the Desktop icon. The instance is a fully operational Centos 7.5 linux image, so you can use it similar to any other desktop. Access the Command Line with SSH For command line access with SSH, you should log in to the public IP with user "centos" and using the keypair you selected when launching the AMI. Both VMD and NAMD can be run this way if no visualization or graphical user interfaces are required. If you are unfamiliar with SSH, please read through Amazon's "Getting Started" Guide. |
Use our Public AMIs If you are a more advanced user and do not wish to go through the Marketplace Listing, you can use any of our available cloud virtual machine images (AMIs) by searching for the listed AMI ID code on Amazon's publicly available AMI list when launching a new instance from the EC2 dashboard. See the EC2 documentation on finding shared AMIs using an AMI ID here.
|
Resources
Recommended Reading Before Getting StartedIn order to access our EC2 virtual machine images, you will need to sign up for an AWS account. Be sure to read through Amazon's "Getting Started" Guide and the EC2 documentation before you begin. For NAMD specific help, please read the NAMD User's Guide and NAMD Mailing List. For VMD specific help, please read the VMD User's Guide and VMD Mailing List.
|
|
Publications |
Presentations
|