NAMD, recipient of a 2002 Gordon Bell Award, a 2012 Sidney Fernbach Award, and a 2020 Gordon Bell Prize, is a parallel molecular dynamics code designed for high-performance simulation of large biomolecular systems. Based on Charm++ parallel objects, NAMD scales to hundreds of cores for typical simulations and beyond 500,000 cores for the largest simulations. NAMD uses the popular molecular graphics program VMD for simulation setup and trajectory analysis, but is also file-compatible with AMBER, CHARMM, and X-PLOR. NAMD is distributed free of charge with source code. You can build NAMD yourself or download binaries for a wide variety of platforms. Our tutorials show you how to use NAMD and VMD for biomolecular modeling.
Breaking News
| NAMD 3.0 New Features - webpage is posted |
| NAMD GPU-resident benchmarks - results and data sets with GPU-optimized configuration posted |
| NAMD 3.0b6 Release - fixes important bugs from last two beta releases |
| NAMD 2.15 ALPHA Release providing GPU-offload support for Intel GPU Max Series. This source code release available on the download page includes SYCL code that can be built using the Intel oneAPI toolkits. Following the download link reveals a page with detailed build instructions. |
Spotlight: Engineering Atomic Detail (Nov 2012)
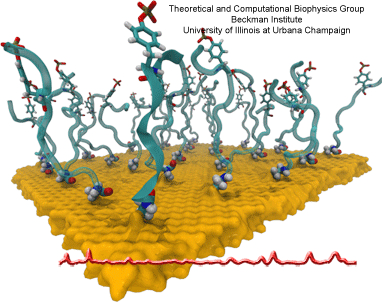
Nanoengineering permits the manufacturing of sensors of unprecedented accuracy to detect biomolecules at very low concentrations as they arise, for example, as signals in living cells. In an important type of cellular signaling, proteins are modified through addition of a phosphate group by other proteins, so-called kinases. Kinases are involved in various types of cancers; therefore, bioengineers seek to develop a nanosensor to detect kinase activity. As described in a recent report, they grafted short peptides, containing a tyrosine amino acid, on nanometer scale gold surfaces. Phosphate groups are negatively charged, and as the groups are transferred from the kinases to the peptide’s tyrosine, the overall charge of the grafted peptides increase. Bioengineers detected then the phosphorylated peptides by applying electrical fields that would drive the charged phosphate group towards the surface or away from it, depending on the voltage polarity; the resulting conformational change of peptides can be recognized by shining light on the nanosensors as optical properties of molecules near metal surfaces are amplified. In order to make the nanodevice really work, the bioengineers needed to optimize the peptide sequence, know how phosphorylation and voltages alter the near-surface conformation of the peptides and how to interpret the measured optical signals. In other words, they needed a microscopic view of the nanodevice! Such view was achieved through molecular dynamics simulations using NAMD and VMD following in the footsteps of similar earlier uses of such simulations as a computational microscope (see Diet and DNA, Sep 2011; Bumpy DNA, Feb 2009; Stretchable DNA, Nov 2005). The combination of nanoengineering and molecular dynamics simulations produced indeed a satisfactory kinase sensor prototype. For more information, visit our kinase sensor website.
Overview
Why NAMD? (in pictures)
How to Cite NAMD
Features and Capabilities
Performance Benchmarks
Publications and
Citations
Credits and Development Team
Availability
Read the License
Download NAMD Binaries
(also VMD)
Build from Source Code
- Git access now available
Run at NCSA, SDSC, NICS, or Texas
Training
NAMD Developer Workshop in Urbana (August 19-20, 2019)
PRACE School on HPC for Life Sciences (June 10-13, 2019)
"Hands-On" Workshop in Pittsburgh (May 13-17, 2019)
Charm++ Workshop in Urbana (May 1-2, 2019)
Enhanced Sampling and Free-Energy Workshop (Sept 10-14, 2018)
NAMD Developer Workshop in Urbana (June 11-12, 2018)
"Hands-On" Workshop in Pittsburgh (May 21-25, 2018)
"Hands-On" QM/MM Simulation Workshop (April 5-7, 2018)
Older "Hands-On" Workshops
Support
Mailing List Issues for Yahoo.com Addresses
Announcements
NAMD 3.0b6 Release (Feb 2024)
NAMD 3.0 New Features (Feb 2024)

NAMD 2.14 Bug Fixes (Apr 2022)
NAMD 2.14 Release (Aug 2020)
NAMD 2.14 New Features
One-click NAMD/VMD in the cloud
QM/MM Interface to MOPAC and ORCA
QwikMD GUI Released in VMD 1.9.3
Previous Announcements
Documentation
Related Codes, Scripts, and Examples
NAMD Wiki (Recent Changes)
Older Documentation
News
AMBER force field use in NAMD for large scale simulation

NAMD GPU-resident benchmarks available

NAMD and VMD share in COVID-19 Gordon Bell Special Prize
NAMD reference paper published online
Coronavirus Simulations by U. Delaware Team
Coronavirus Simulations on Frontera Supercomputer
Breakthrough Flu Simulations
Oak Ridge Exascale Readiness Program
Prepping for Next-Generation Cray at NERSC
Supercomputing HIV-1 Replication
How GPUs help in the fight against staph infections
Computational Microscope Gets Subatomic Resolution
Opening New Frontiers in the Battle Against HIV/AIDS
HIV Capsid Interacting with Environment
Assembling Life's Molecular Motor
Older News Items