TCBG and the National Strategic Computing Initiative
31 August 2015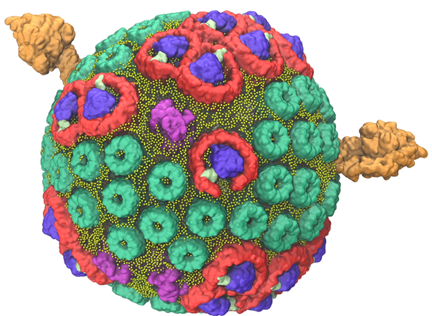
TCBG has already simulated this autonomously functioning organelle, the photosynthetic chromatophore, a first step on the way to whole-cell simulation.
"As leaders in scientific computing in America, we were really getting very, very nervous," says Klaus Schulten, "And now suddenly we are getting this loud directive from the president, that all the government agencies should make this their priority." Schulten was worried about the future of supercomputing in the United States, but on 29 July 2015, Schulten's frustration turned into elation when President Barack Obama issued an executive order to "maximize the benefits of high-performance computing" in the country. The President's executive order basically means that building the next generation of supercomputers, so called "exascale" machines, is now a national priority. And Klaus Schulten knows exactly what this could mean for him in the field of computational biophysics.
In fact, even before Obama made his announcement, Schulten was contacted by the director of the University of Illinois supercomputer center (the NCSA) and asked the general question: What could you do with a supercomputer one hundred times more powerful than Blue Waters? (The Blue Waters supercomputer at NCSA is a 10 petaflop machine). The NCSA was soliciting answers to this question to help facilitate the eventual executive order. Schulten immediately wrote a response to the NCSA, which is included below.
But why would Schulten's group, the TCBG, be so excited about the possibility of an exascale computer? Well, the intersection of TCBG and the supercomputer comes into play because living cells are made of molecules. The TCBG has been looking at the molecular and atomic world since its founding in 1989, very often utilizing its tools NAMD, VMD, and MDFF in concert with high-performance computing. In recent years, TCBG has been able to examine bigger and bigger assemblies of molecules, with the goal of explaining biological organization. An exascale computer offers one hundred times more power than is currently available and hence could allow TCBG to simulate a system as big as 0.10 μm3, which is about one hundred times larger than is presently feasible. This is close to the size of a small cell.
In general, computers are playing a larger and larger role in society. And Schulten says the use of computers in the health sciences and health treatment fields is no exception. Schulten expects supercomputers can play a larger role in these fields, especially now that federal agencies are directed to work together to bring an exascale supercomputer online within the next decade. Continue reading below to see what Schulten believes will be possible with an exascale machine in the field of computational biophysics, and how this can translate to the world of health.
Computational Microscopy of Whole Living Cells, A Window to Fundamental Medical Knowledge
By Klaus SchultenThe next generation of computers can complete the dynamic picture of the fundamental living system, the cell, integrating a century of discoveries in the Sciences into the ultimate answer of the age old question: What is life? Computational biologists, in close partnership with experimental biologists, exploited three decades of increasing computer power at NSF Supercomputer Centers to describe at the atomic level single proteins (1000 atoms), proteins in their cell environment (100,000 atoms), complexes of proteins like a virus shell (1 million atoms), and, finally, on Blue Waters even fundamental compartments in living cells, namely a cell's autonomously functioning organelles (billion atoms).
The HIV capsid. This structure was determined with the method Molecular Dynamics Flexible Fitting.
The cell-scale computations that I described are data driven (X-ray crystallography, cryo-electron microscopy, and other methods). My group has developed a hugely successful method called "Molecular Dynamics Flexible Fitting" (MDFF) that establishes the huge macromolecular structures found in living cells and of whole viruses driven by the data. The method is only six years old, yet it had produced already many structures, the most famous one being the structure of the HIV virus capsid. The data driven simulations require often only 10-100 milliseconds, but in some cases longer simulations are needed. Such longer simulations can be effectively achieved through advanced sampling algorithms that can reach millisecond time scales.
So in short, simulations of very large biomolecular assemblies alluded to above are prime examples for data-driven computation. Together with modern statistical sampling techniques, exascale computing–the goal set by the President's National Strategic Computing Initiative directive–will permit researchers to cover biologically relevant time scales for large and exciting biomolecular systems and eventually simulate for the first time a whole living cell.
More Information
- See this History of MDFF, which includes the story of the HIV capsid structure.
- See this story, Researchers Build Bacteria's Photosynthetic Engine, which discusses TCBG efforts to build the structure of the photosynthetic chromatophore on the Titan supercomputer. The chromatophore contains over 100 million atoms, and the ensuing Titan simulations are a key milestone on the way to whole-cell simulation.



