Prostaglandin H2 synthase-1
Structure
Eicosanoid compounds such as prostaglandins are formed from the essential fatty acid, arachidonic acid (AA), by the enzymes in the arachidonate cascade. The enzyme prostaglandin H2 synthase-1 (PGHS-1) catalyses the first committed step in the biosynthesis of prostaglandins, converting AA to prostaglandin H2 [1]. Non-steroidal anti-inflammatory drugs like aspirin, ibuprofen, and indomethacin directly target PGHS-1, and inhibit the conversion of AA [2]. Prostaglandins initiate and modulate cell and tissue responses involved in many physiological processes such as platelet aggregation and inflammation.
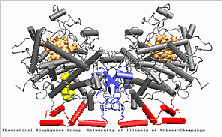
PGHS-1 is an integral membrane protein located primarily in the endoplasmatic reticulum [1]. The enzyme is bifunctional and catalyses a cyclooxygenase and a peroxidase reaction. PGHS-1 consists of two identical monomers related by a non-crystallographic two-fold symmetry axis. The amino-terminal residues form a small compact domain, held together by intra-domain disulphide bonds, similar to the structure of the epidermal growth factor (EGF). The highly amphiphatic membrane binding motif inserts the enzyme into one leaflet of the lipid bilayer. The globular structure of the catalytic domain contains the cyclooxygenase and peroxidase active sites. The substrate of PGHS-1 (arachidonic acid) is depicted in the cyclooxygenase site.
Simulations
Molecular Dynamics simulations with external forces (SMD and TMD [3]) are employed to study the unbinding and binding of fatty acids in the cyclooxygenase (COX) site of Prostaglandin H2 Synthase-1 [4]. Simulations with the natural COX substrate, arachidonic acid (AA), inside the COX binding channel reveal sequences of concerted bond rotations in the fatty acid alkyl chain which obviate the need for gross conformational changes in the protein and substrate during unbinding and binding. The all-cis structure of AA, with double bonds separated by two single bonds, facilitates easy access to the COX channel and correct positioning inside the active site for the COX chemistry to occur. Two derivatives of AA, one with a cis double bond changed to a trans configuration and the other with a double bond reduced to a single bond, are also studied. In both cases the concertedness of bond rotations in the fatty acid chain is diminished and larger forces are required to move the fatty acid inside the COX channel. While no large scale conformational changes in the protein are observed, important motions of residues near the mouth of the COX channel are found and analyzed. In particular, a conformational ``switch`` involving Arg83, Glu523 and Arg120 is seen to mediate the movement of the substrate from the membrane to the channel.
Trajectories
| System | SMD unbinding | TMD binding |
|---|---|---|
| Arachidonic Acid (AA) unbinding: 250 ps binding: 70 ps |
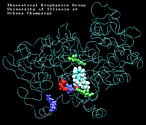 (2.2MB) |
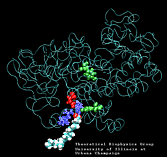 (1.4MB) |
| "8-trans" AA unbinding: 250 ps binding: 70 ps |
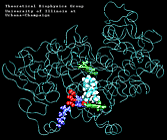 (2.8MB) |
 (1.3MB) |
| "8-single" AA unbinding: 250 ps |
 (2.6MB) |
|
| AA with water present at the entrance of the binding pocket unbinding: 620 ps |
 (3.0MB) |
|
Personnel
References
- D. Picot, P.J. Loll, and M. Garavito. The X-ray crystal structure of the membrane protein Prostaglandin H2 synthase-1. Nature 367:234-249, 1994.
- W.L. Smith and D.L. DeWitt. Biochemistry of Prostaglandin H Synthase-1 and Synthase-2 and Their Differential Susceptibility to Nonsteroidal Anti-Inflammatory Drugs. Seminars in Nephrology 3:179-194, 1995.
- J. Schlitter, M. Engels, P. Krueger, E. Jacoby, and A. Wollmer. Targeted Molecular Dynamics Simulation of Conformational Change - Application to the T -< R Transition in Insulin. Molecular Simulation 2-6:291-308, 1993.
-
Publications Database Simulated unbinding and binding of fatty acid substrates in the cyclooxygenase site of prostaglandin H2 synthase-1. Ferenc Molnar, Lawrence S. Norris, and Klaus Schulten. Progress in Reaction Kinetics and Mechanism, 25:263-298, 2000.