More Research Categories
The biomedical sciences are enjoying a wealth of data from innovative new techniques in structure determination, DNA sequencing at the level of entire genomes, and direct manipulation and observation of single molecules. The Theoretical and Computational Biophysics Group complements these efforts through the most advanced molecular modeling, bioinformatic, and computational technologies, extending and refining these technologies in response to the experimental advances. Our research focuses on the modeling of large macromolecular systems in realistic environments. These efforts have produced insight into biomolecular processes coupled to mechanical force, bioelectronic processes in metabolism and vision, and the function and mechanism of membrane proteins.

Exchange of materials and information between a living cell and its environment is mediated and regulated by membrane proteins. These proteins are involved in the regulation of electrical activity of the cell, transport of water and water soluble materials across the membrane, and production of ATP. Membrane receptors are the sites for detection informational signals, such as neurotransmitters and hormones, light, and even mechanical stress. The atomic-resolution structures of a few membrane proteins have been solved, and recent advances in structure determination of membrane proteins promise more structure to be solved soon. Our group studies the mechanism of function of membrane proteins with various computational methodologies. The proteins are simulated in their natural environment, i.e., embedded in fully hydrated patches of lipid bilayers and under constant pressure and temperature conditions. The main objective is to understand how specific structural motifs and/or chemical interactions in a protein play a role in its function.
Many cellular processes involve transformations and sustenance of mechanical forces. Forces arise as input, output and signals in cellular machines, such as ATP synthase, motor proteins, and cell surface receptors. Proteins functionally dealing with mechanical forces belong to the research subjects of the emerging new field mechanobiology. Examples among them are the giant muscle protein titin sustaining forces developed in muscle cells, the extracellular matrix fibronectin forming elastic fibrils connecting cells, mechanosensitive channels being switched on by tension applied to the cell membrane, integrins conveying mechanical signals across membrane, ankyrin amplifying weak force signals for the mechanotransduction process in hearing cells, lac repressor grappling with the regulated DNA, Fo-ATPase converting cell's electrical energy into rotation, and various ligand-receptor complexes. What is the physical mechanism underlying the mechanical functions of these proteins? Over the past decade the group has developed and applied a novel computational approach termed steered molecular dynamics to study the structure-function relationship of these mechanical proteins.
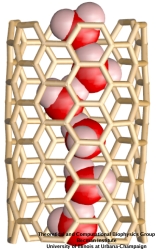
Molecular modeling provides nanoscale images at atomic and even electronic resolution, predicts the nanoscale interaction of yet unfamiliar combinations of biological and inorganic materials, and can evaluate strategies for redesigning biopolymers for nanotechnological uses. The methodology's value has been reviewed for three uses in bionanotechnology. The first involves the use of single-walled carbon nanotubes as biomedical sensors where a computationally efficient, yet accurate description of the influence of biomolecules on nanotube electronic properties and a description of nanotube - biomolecule interactions were developed; this development furnishes the ability to test nanotube electronic properties in realistic biological environments. The second case study involves the use of nanopores manufactured into electronic nanodevices based on silicon compounds for single molecule electrical recording, in particular, for DNA sequencing. Here, modeling combining classical molecular dynamics, material science, and device physics, describes the interaction of biopolymers, e.g., DNA, with silicon nitrate and silicon oxide pores, furnishes accurate dynamic images of pore translocation processes, and predicts signals. The third case involves the development of nanoscale lipid bilayers for the study of embedded membrane proteins and cholesterol. Molecular modeling tested scaffold proteins, redesigned lipoproteins found in mammalian plasma that hold the discoidal membranes in shape, and predicted the assembly as well as final structure of the nanodiscs. In entirely new technological areas like bionanotechnology qualitative concepts, pictures, and suggestions are sorely needed; the three exemplary applications document that molecular modeling can serve a critical role for the new bionanotechnology, even though it may still fall short on quantitative precision.
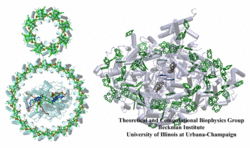
Photosynthetic complexes provide fascinating challenges to biophysicists. With the availability of recent atomic structures for protein-pigment complexes such as photosystem I in cyanobacteria and plants, it is possible to form a comprehensive picture of the light absorption and excitation migration processes based on an atomic level quantum mechanical description. This kind of structural analysis not only provides a rigorous test for our understanding of the physics of these mechanisms through a comparison to spectroscopy and kinetics experiments, but it also forms a framework within which the organizational principles for multi-component pigment-protein assemblies can be investigated.
Knowledge of the mechanism of association, dissociation and unfolding of macromolecules is important for many biological processes. Among the examples are the binding and dissociation of substrates of enzyme reactions, the recognition of ligands by their receptors or the elastic resopnse of mechanical proteins. In order to study such processes external forces can be applied reducing energy barriers and therefore increasing the probability of unlikely events on the time scale of molecular dynamics. This approach has the advantage that it corresponds closely to micromanipulation through atomic force microscopy or optical tweezers. The external force techniques can be applied to study many processes, including dissociation of avidin-biotin complex, dissociation of retinal from bacteriorhodopsin, stretching of titin, etc. The molecular dynamics program NAMD, developed in the group, is capable of performing several different kinds of SMD, including rotation or translation of one or more atoms. The group's molecular graphics program VMD provides a powerful means of visualizing these simulations, and through the Interactive Molecular Dynamics (IMD) interface can even allow SMD simulations to be performed in real time.
Many important biological processes taking place in cells are driven and controlled by events that involve electronic degrees of freedom and, therefore, require a quantum mechanical description. An important example are enzymatically catalyzed, cellular biochemical reactions. Here, bond breaking and bond formation events are intimately tied to changes in the electronic degrees of freedom. Key events during photosynthesis in plants and energy metabolism in eucaryotes also warrant a quantum mechanical description - from the absorption of light in the form of photons by the photosynthetic apparatus to electron transfer processes sustaining the electrochemical membrane potential. Because of the importance of sensing light to both plants (for regulating vital functions) and animals (for vision), the interaction between light and biological photoreceptors is widespread in nature, and also requires a quantum mechanical description. A prime example is the protein rhodopsin which is present in the retina of the human eye and plays a key role in vision. Our computational tool are combined quantum mechanical/molecular (QM/MM) simulations, that allow to combine an electronic level description of the active region with a classical model of the environment provided by the remainder of the biomolecular system and solvent. This allows us to study the electronic level processes underlying these systems in their natural cellular environment.

Concerned with both central and peripheral nervous system in living organisms, the field of neurobiology includes explorations into the structure, function, development, biochemistry, physiology and pathology of the nervous system, with the level of analysis ranging from molecular through cellular levels of analysis. At the Theoretical and Computational Biophysics Group, research topics have included the use of neural networks to control motor movement, including visuo-motor control (based on models of the visual cortex and studies of its development) and applicability of neural network concepts to control robotic arms.
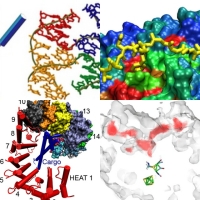
The nature of modern science is that it is ever-changing, energetically crossing boundaries heretofore defined by traditional areas of inquiry. Research at the Theoretical and Computational Biophysics group reflects this dynamic, with studies employing theoretical perspectives and methodological approaches or addressing topics that don't fall easily into one of the above categories. Included in this broad category are studies of a four-way DNA junction, the nuclear pore complex, gas transport in hydrogenase that may provide a source of renewable fuel, and other topics.
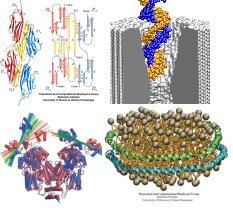
The Theoretical and Computational Biophysics group works side by side with experimental collaborators to explore a variety of seminal research topics, such as the structure of aquaporin, the agregation of lipoproteins, the binding of the lac repressor protein to DNA, the role of the titin-telethonin complex in muscle fiber, and the translocation of DNA through nanopores. Through our collaborative projects, we are able to simulate and make predictions about systems which are of great interest to experimentalists and the scientific community at large, and can fully utilize the critical interplay between theory and experiment.



