Collaboration and Service Projects
Recent advances in experimental techniques such as cryo-electron microscopy (cryo-EM), nuclear magnetic resonance (NMR) spectroscopy and X-ray crystallography are pushing the limits for characterizing macromolecular complexes in terms of size and detail. While each technique unveils critical aspects of molecular and cellular architecture, the potential for a single experimental method to produce a comprehensive, all-atom description of a biomolecular system in its native state remains limited. To overcome this limitation, computational molecular modeling techniques, such as molecular dynamics (MD) simulations, have been playing a crucial role, addressing structural details and dynamic events at the molecular level which are otherwise inaccessible. The NIH Center for Macromolecular Modeling and Bioinformatics supports the biomedical research community by engaging in Collaboration and Service (C&S) projects that leverages Center technology, expertise and facilities to bridge experiments and computation. Here we give a representative sample of the over 40 C&S projects that are currently conducted by the Center.
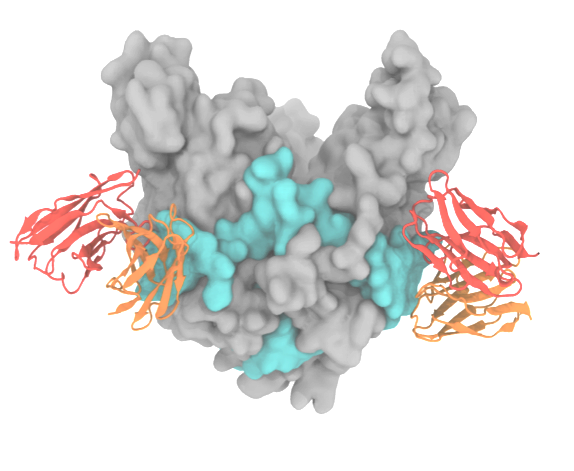
Cell entry is the first stage of the viral infection cycle. Enveloped viruses such as influenza A and Ebola employ glycoproteins (GP), located on the viral surface, to facilitate cell entry and exit. Inhibiting the viral GP by using drugs or antibodies is therefore an effective way to impair the virus's ability to enter the cell. In particular, antibodies that bind with high affinity and specificity are valuable as they can be used for diagnostic and therapeutic purposes. Despite its biomedical importance, developing novel antibodies using solely experimental methods remains a time-consuming and expensive process. Center-developed technologies are employed to improve a computational antibody design method that targets viral GP.
Read more about the Center's virus research and the the Center's five year research plan on designing antibodies.
Collaborating Investigators: C.D. Maranas (Penn. St. U.); P. Tessier (RPI); B.A. Seaton (Boston U.)
Funding: NSF CBET-1133040, 07/2012--06/2016 (CDM); NSF CBET-1159943, 09/2012--08/2016 (PT); NSF ACI-1524703, 07/2015--06/2016 (KS)
Key Publication: Goh et al., Biochemistry, 2016
Adverse drug-drug interactions (DDI) are a significant problem in clinical practice, especially for patients taking multiple drugs on a regular basis. A major type of DDI involves the interference of different drugs at the level of metabolism, and is observed when the metabolism of one drug is affected by the presence of co-administered drugs. In the human body, 70% of drugs are metabolized by cytochromes P450 (CYPs), a large family of enzymes involved in the biotransformation of drugs. Hence, drug metabolism mediated by CYPs constitutes a major determinant in the occurrence of several adverse DDI. Therefore, a detailed description of the molecular mechanism underlying DDI mediated by CYPs will provide medicinal chemistry and pharmacology with a way to prevent adverse drug effects in clinical practice.
Collaborating Investigators: S.G. Sligar (UIUC); A. Das (UIUC)
Funding: NIH R01-GM101048, 06/2012-06/2016 (SGS,ET); AHA 15SDG25760064, 01/2015-01/2018 (AD)
Read more about the Center's five year research plan on drug interactions in Cytochrome P450.
Key Publication: Denisov et al., Biochemistry, 2015
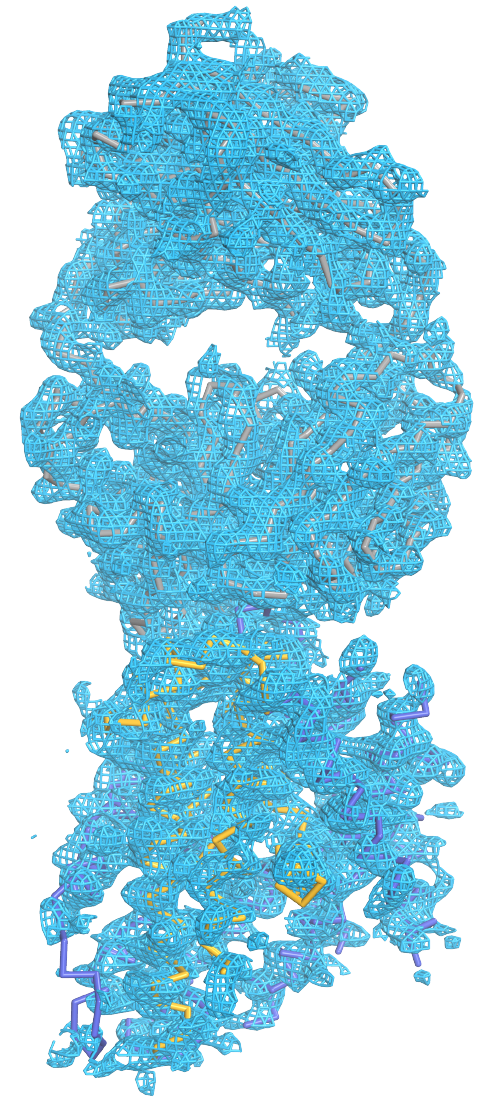
Protein degradation is vital for a variety of essential cellular processes, including protein quality control, cell cycle regulation, adaptive immune response, and apoptosis. The 26S proteasome is responsible for the vast majority of regulated intracellular protein degradation and an important drug target for multiple diseases, including cancer, neurodegenerative diseases, and immuno-inflammatory disorders. The 26S proteasome is an ATP hydrolysis driven 2.5 MDa molecular machine that recruits, unfolds, and degrades poly-ubiquitin tagged proteins through a complex interaction clockwork of over 60 known protein subunits. Despite its substantial role in the cell's life cycle, the proteasome's detailed atomic structure and mechanism still remain elusive. Center-developed software tools will be employed to investigate the underlying molecular mechanism of (1) substrate recruitment and (2) substrate unfolding by the proteasome .
Read more about the Center's proteasome research and the Center's five year research plan to investigate protein degradation.
Collaborating Investigators: W. Baumeister (MPI Munich)
Funding: NSF: PFC: CPLC: PHY1430124, 01/2004 - 08/2019 (KS); DFG: CIPSM Excellence Cluster (EXC 114), 06/2012 - 06/2017 (WB); DFG: CRC 1035, 06/2012 - 06/2017 (WB)
Key Publication: Schweitzer et al., PNAS, 2016
For most membrane proteins, the process of insertion and integration into lipid membranes takes place concurrent with their translation and synthesis by the ribosome. Given their therapeutic relevance, the insertion pathways of nascent trans-membrane proteins into lipid membranes deserve more attention than they have hitherto received. The ribosomal insertion machinery consists mainly of YidC, SecY and the holo-translocon supercomplex; the holo-translocon supercomplex itself is composed of YidC, SecY and SecDF. Depending on size and nature of the inserting nascent protein, the membrane insertion process is performed by either YidC or SecY alone, or by both of them working in concert within the holo-translocon supercomplex. Here, we plan to (1) model an all-atom structure of a membrane-embedded ribosome-bound holo-translocon supercomplex , and then (2) investigate how conformational changes of the holo-translocon supercomplex regulates membrane proteins insertion pathways.
Read more about the Center's ribosome research and the Center's five year research plan for the ribosomal holo-translocon supercomplex.
Collaborating Investigators: C. Schaffitzel (U. of Bristol & EMBL)
Funding: ERC (Starting Grant), 08/2012-09/2017 (CS); NIH 2R01GM067887-12, 06/2016-06/2019 (KS)
Key Publication: Wickles et al., eLife, 2014
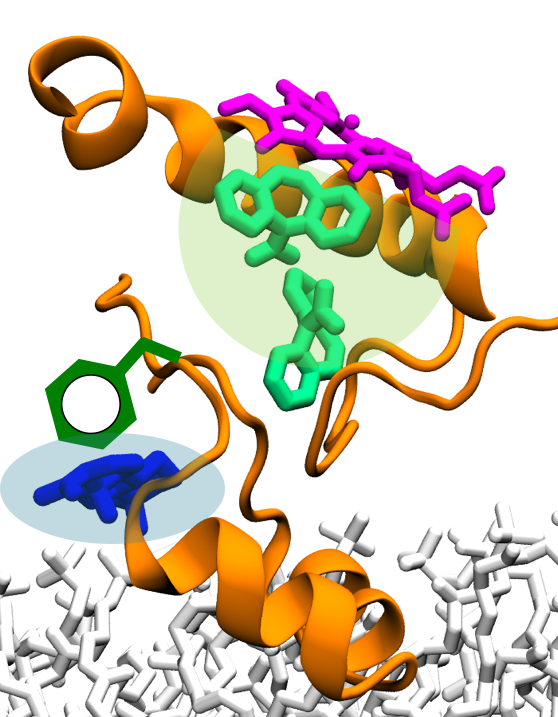
The ubiquitous Nramp family of transport proteins, a member of the LeuT family, facilitates acquisition of metal ions, including the absorption of dietary iron in mammals. Nramp proteins have an essential role in host defense against pathogens. Thus, known Nramp mutations lead to impaired immune response. Furthermore, since Nramp is also crucial for iron uptake into the cytosol, its mutations cause anemia in humans and rodents. Here, tools developed at the Center will be applied to resolve (1) how Nramp selectively binds and transports essential transition metal ions (Fe2+, Co2+, Mn2+, and Zn2+) over toxic metals (Cd2+ and Pb2+) or the more abundant alkaline earth metals (Ca2+ and Mg2+) , and (2) why Nramp mutants fail to achieve such selectivity for transition metals.
Read more about the Center's five year research plan for metal transporters.
Collaborating Investigators: R. Gaudet (Harvard U.)
Funding: NIH 1R01GM120996-01, 02/2016-01/2021 (RG); NIH 2R01GM067887-12, 06/2016-06/2019 (KS)
Key Publication: Bozzi et al., PNAS, 2016
Nanosensors utilizing nanoscale holes in solid-state membranes, known as nanopores, are exten- sively explored as an analytical tool for accurate, fast, and inexpensive human genome sequencing and epigenome mapping. In order to enhance the sensitivity of nanopore-based devices, a variety of approaches have been proposed to control the motion of molecules while they move through nanopores, including the use of multilayered solid-state membranes containing motion-control layers aside from a central sensing layer. Here, Center-developed technologies will enable: (1) design of the multilayered nanopore devices for precise control over molecular motion, and (2) optimization of the nanopore devices for accurate recognition and positioning of DNA nucleotides.
Read more about the Center's nanosensors research and the Center's five year research plan for nanopore-based sensors.
Collaborating Investigators: J.-P. Leburton (UIUC); R. Bashir (UIUC); A. Radenovic (EPFL)
Funding: Beckman Seeding Grant, 05/2014 - 05/2016 (JPL,RB,KS); Oxford Nanopore Grant, 06/2012 - 06/2017 (JPL,RB); SNSF-ERC Consolidator Grant BSCGI0 157802, 06/2015 - 06/2020 (AR); Swiss NSF 200021 153653, 03/2014 - 03/2017 (AR)
Key Publication: Hu et al., Nano Letters, 2015
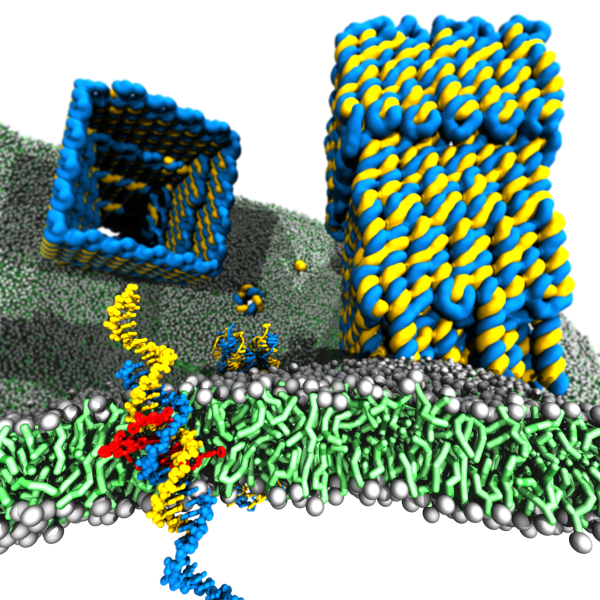
Membrane protein channels involved in cellular signal transductions are fascinating biological sensors with high selectivity and efficiency. Recently, DNA origami nanostructures emerged as highly customizable mimetics of biological membrane channels. A typical DNA membrane channel is assembled from several DNA double helices arranged into a polygon pattern, with the central cavity forming the transmembrane pore. To facilitate insertion of the DNA channel into a lipid bilayer membrane, the DNA helices are chemically modified to carry hydrophobic anchors. Until now, most of the DNA channels featured four or six DNA helices arranged in a square or a hexagon, with the inner channel diameter varying between 1 and 2.5 nm. The outstanding problems of the field are: (1) Establishing the relationship between the design of a DNA channel and its function as a passage across the lipid bilayer membrane. (2) Improving the designs of DNA channels to realize selective transmembrane transport controlled by external stimuli. (3) Engineering DNA channels for applications in sensing, drug delivery and artificial tissues technologies.
Read more about the Center's five year research plan for DNA membrane channels.
Collaborating Investigators: U. F. Keyser (U. of Cambridge)
Funding: NSF ECC-1227034, 09/12-08/17 (AA); DMR-1507985, 08/15-07/18 (AA); ERC-2014-CoG (Designer- Pores), 07/15-07/20 (UFK)
Key Publication: Yoo et al., Journal of Physical Chemistry Letters, 2015
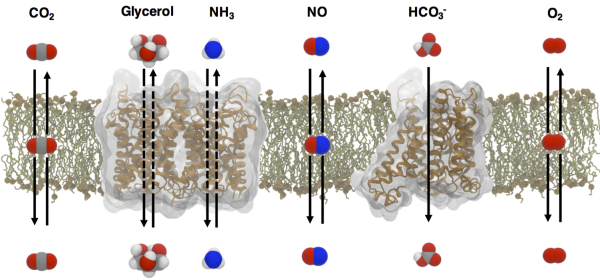
Small gases, such as dioxygen (O2), nitric oxide (NO), carbon dioxide (CO2) and ammonia (NH3), participate in numerous biochemical processes in living cells. CO2 and NH3 are involved in acid-base homeostasis; NO is involved in signaling; and O2 is involved in metabolism. As a vast majority of these processes take place inside of cells, gas transport across biological membranes becomes one of the most fundamentally physiological processes. The movement of polar NH3 gases is facilitated through membrane channels, such as Rh proteins and aquaporins (AQPs). Nonpolar O2, NO and CO2 gases may simply diffuse through the lipid bilayer, but there are membranes that are impermeable to these small gases. AQPs are one of the most extensively studied membrane channels and are comprised of thirteen isoforms in human cells with unique selectivity features to gases. Not only are they known to function as water channels, they also faciliate the transport of glycerols and NH3, as well as CO2 and NO. Although gas permeation across biological membranes has been extensively studied in particular by Boron, a long-time collaborator, the existence of gas channels has remained controversial especially for CO2. Thus, fundamental aims to be addressed include: (1) elucidating the structural mechanism underlying solute selectivity in AQPs, and (2) the role of AQPs in transporting gases across membranes.
Read more about the Center's five year research plan for gas transport through biological membranes.
Collaborating Investigators: W. F. Boron (Case Western Reserve U.)
Funding: NIH U01-GM111251, 01/2015-12/2019 (WFB); DOD ONR N00014-1512060, 02/2015-02/2018 (WFB); DOD ONR MURI-2016, 06/2016-05/2021 (WFB)
Key Publication: Geyer et al., American Journal of Physiology - Renal Physiology, 2013
The ability of proteins to fold into their native state is essential for cell function; misfolded proteins not only lose their function, but can also cause neurodegenerative diseases, including Alzheimer and Huntington. Study of protein folding can aid in preventing protein misfolding diseases and in designing proteins with novel functions. Although most cellular proteins fold on timescales of milliseconds to seconds, the current study will focus on a 80-residue model protein named λ-repressor, which has been experimentally characterized to fold on a timescale of less than 20 μs. Our collaborator Gruebele has developed several mutants of λ-repressor that can fold on different time scales. The project will employ Center-developed long-time MD simulation with NAMD to investigate: (1) the folding pathway of λ-repressor and (2) the effect of mutations on folding of the λ-repressor.
Read more about all the Center's protein folding research and its five year research plan.
Collaborating Investigators: M. Gruebele (UIUC)
Funding: NIH R01 GM093318, 05/2010 - 04/2017 (MG)
Key Publication: Liu et al., JACS, 2014
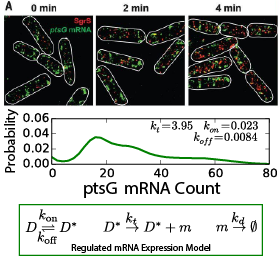
A high level of gene regulation is achieved by regulatory RNAs that can modulate mRNA translation independently from transcriptional signals. This principle is not only crucial to complete the picture of gene regulation; it also forms the basis of small interfering RNA therapies that are under development to treat a host of diseases, including several types of cancer. Variants of regulatory RNA exist in all domains of life and are known in bacteria as small RNA (sRNA). Seminal single-molecule imaging studies have yielded insight into the in vivo action of the E. coli sRNA SgrS (sugar-transport related sRNA), which regulates sugar uptake by its selective recognition and the thereby triggered degradation of the mRNAs for ptsG and manX (which encode the primary glucose and mannose transporters in E. coli). In this collaboration, Center-developed whole-cell simulation technologies incorporated in Lattice Microbes (LM) are used to investigate (1) the role of spatial localization on SgrS-mRNA binding kinetics and competition and (2) the downstream effects of SgrS control of sugar uptake on cellular metabolism.
Read more about our Center's five year research plan for small RNA regulation in E. coli.
Collaborating Investigators: T. Ha (John Hopkins U.); J. Fei (U. of Chicago); C. Vanderpool (UIUC)
Funding: NIH R01-GM112659, 01/2015 - 11/2019 (TH, ZLS); NSF: PFC: CPLC: PHY1430124, 08/2014 - 08/2019 (TH, JF, ZLS)
Key Publication: Peterson et al., PNAS, 2015