Mechanism of Hexameric Molecular Motors
Understanding structure-function relationship of molecular motors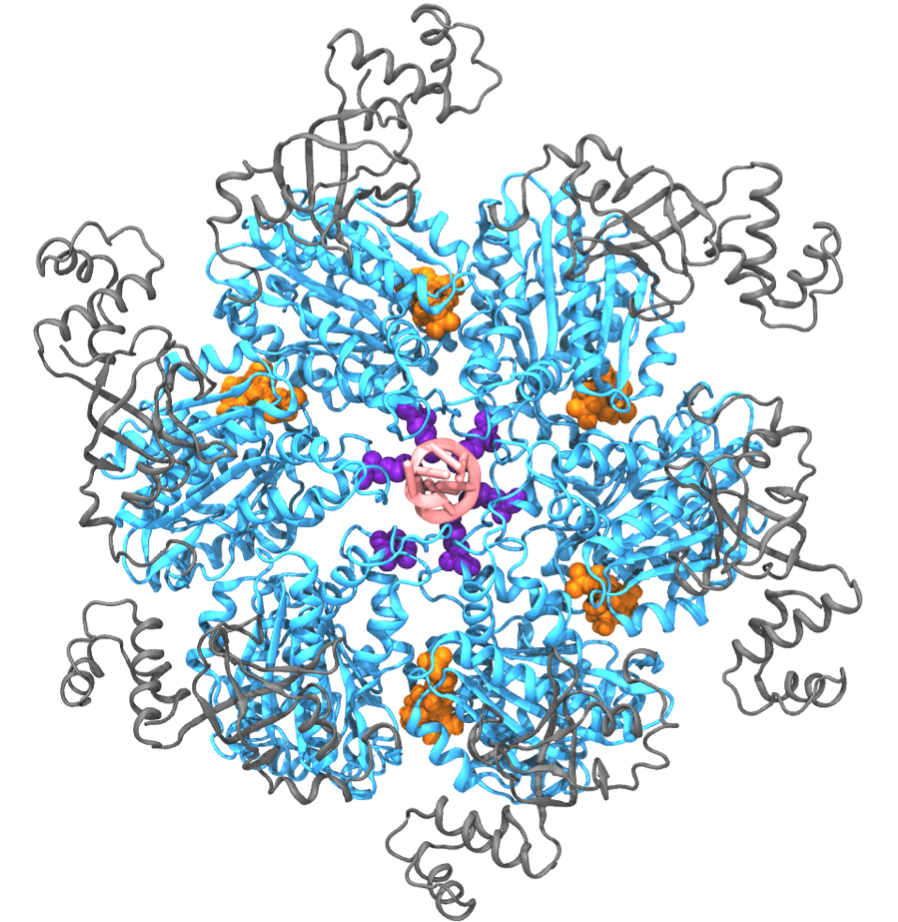
Fig. 1: Exemplary hexameric motor (Rho, pdb code: 3ICE). Hexameric molecular motors are ring-shaped, multi-subunit enzymes that convert the energy from ATP hydrolysis to the generation of force or torque, by which the motors perform mechanical work. These molecular motors participate in many cellular processes, including DNA repair, gene expression, protein degradation, and maintenance of pH homeostasis [ Lyubimov et. al., COSB, 2011 ]. Despite intense research efforts, the atomic-level mechanism transmitting chemical energy from hydrolysis into mechanical force is still unclear. Molecular dynamics (MD) simulations, though capable of providing atomic details, are limited in the study of molecular motors owing to the challenge that these motors function on a millisecond time scale which, for a long time, could not be covered computationally. Employing novel computational methods, namely so-called pathway sampling techniques, we are now able to study the millisecond dynamics of an exemplary hexameric motor system, namely Rho helicase (Fig. 1). Our results reveal unprecedented atomic level details of the motor action mechanism that agree well with existing experimental results. The new approach promises also an opportunity to resolve the mechanism of other molecular motors. We have reported the study on Rho in [ Ma and Schulten, JACS, 2015 ]. See our highlight here . |
Rotary reaction mechanism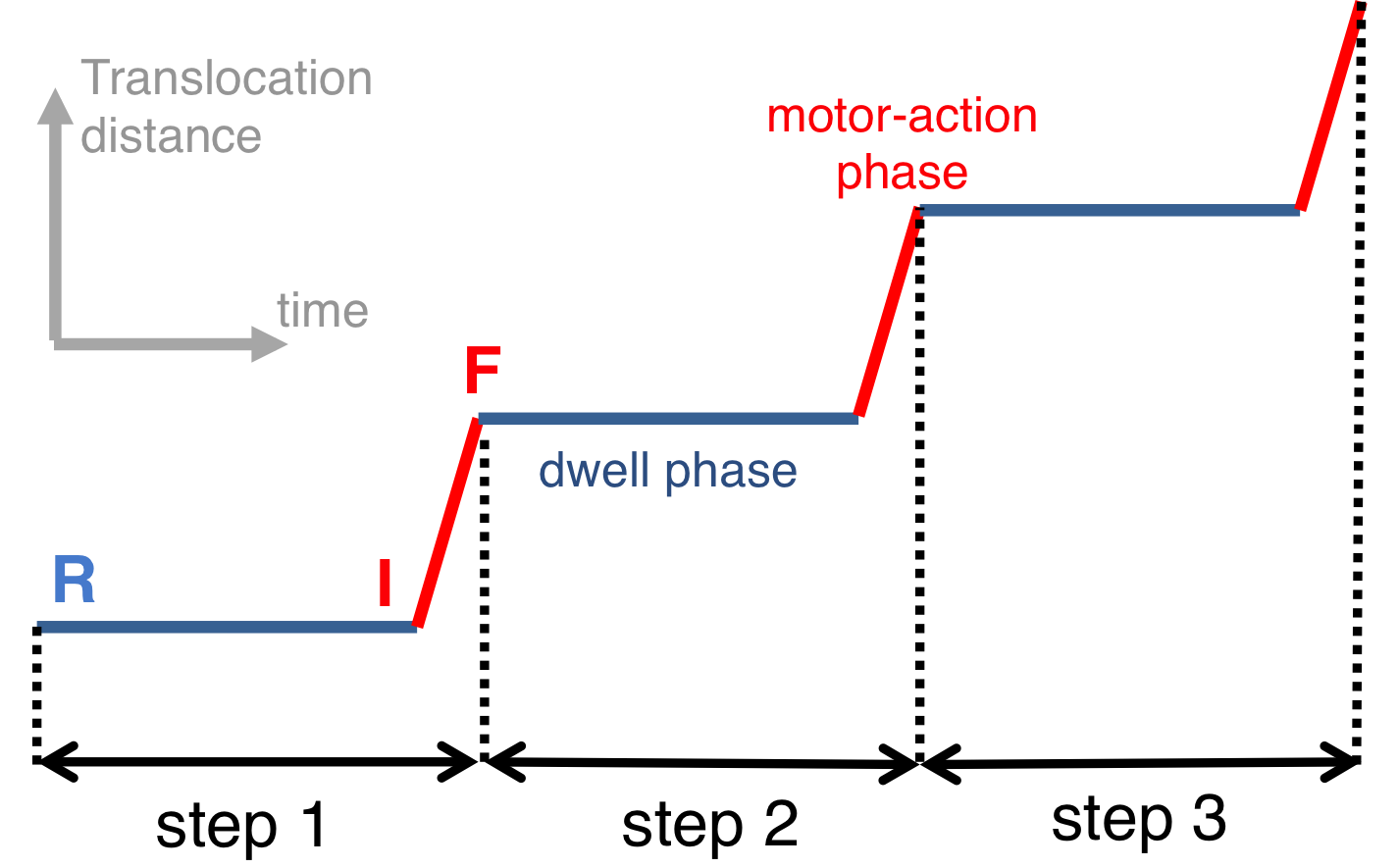
Fig. 2: Schematic illustration of dwell and motor-action phases of a hexameric motor. 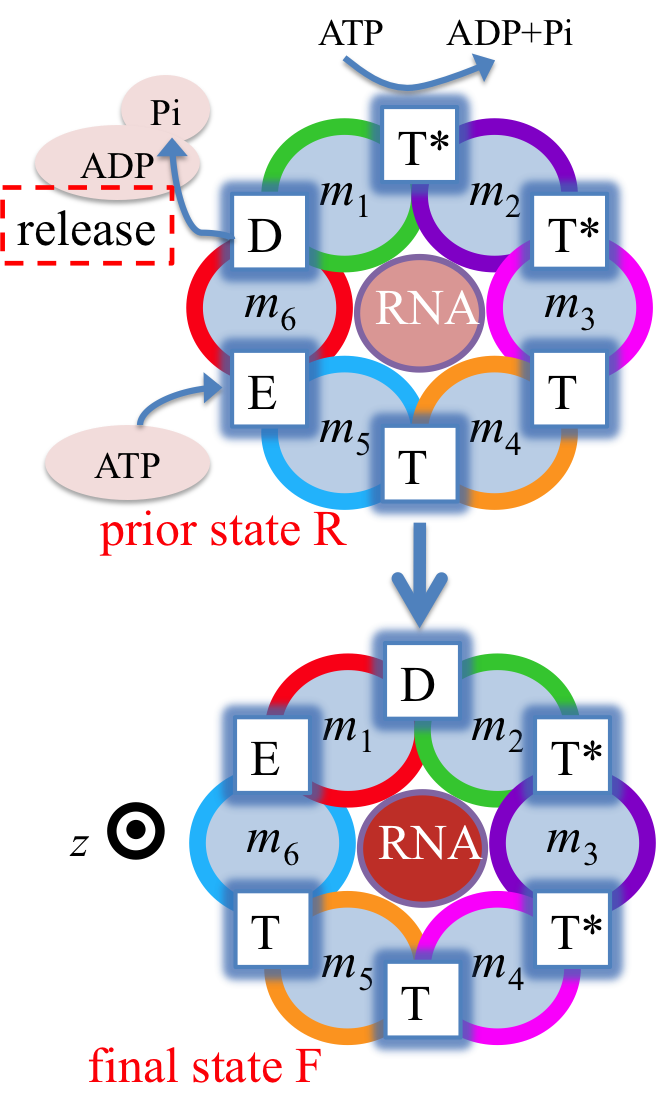
Fig. 3: Proposed rotary reaction mechanism of Rho. Active sites at the interfaces are labeled according to the state of ligands: hydrolysis-competent (T*), ATP-bound (T), "old" product (D), and empty (E) state. The function of the ring-shaped motors (such as Rho) is carried out in a series of repeated steps (e.g. steps 1, 2, 3 in Fig. 2), by generating linear force or torque on the substrate (nucleic acid strand, polypeptide, or central stalk) inside the central pore of the ring. Each step of the motors contains a dwell phase of relative long duration (colored blue in Fig. 2) and a motor-action phase (colored red in Fig. 2). During the dwell phase, the motor becomes energetically charged through the occurrence of three chemical processes, namely (1) ATP binding, (2) release of existing (i.e., "old") ATP hydrolysis product, and (3) hydrolysis of ATP into "new" product, before the next motor-action phase takes place. Interestingly, none of the three chemical processes during the dwell phase are directly involved in mechanical force generation since these processes happen at subunit-subunit interfaces along the ring, distant from the substrate binding site inside the central pore where mechanical force is actually being applied. Rho is a key factor in bacterial gene expression and regulation, with a primary role in transcription termination through translocating the nascent mRNA and terminating transcription upon reaching the RNA polymerase. The asymmetric structure of Rho [ Thomsen and Berger, Cell, 2009 ] reveals that the six interfaces between Rho's six subunits exhibit a specific circular pattern of different conformational states in the ATP hydrolysis cycle, the states being related to six sequential ligand binding states T*, T*, T, T, E, D shown and defined in Fig. 3 (prior state R). In binding ATP, releasing ADP + Pi and hydrolyzing ATP to ADP, the pattern of ligand binding states in the prior state R, T*T*TTED, shifts to DT*T*TTE in the final state F. The ligand binding states of R, after a 60° clockwise rotation around the z-axis, are the same as those of F. The different colors of the six subunits' circular peripheries indicate schematically that the subunits assume different interface conformations. One should note that the conformational pattern also shifts 60° around the z-axis from R to F; however, the whole hexamer itself does not rotate. |
Characterizing millisecond dynamics of hexameric helicase Rho in atomic-level detail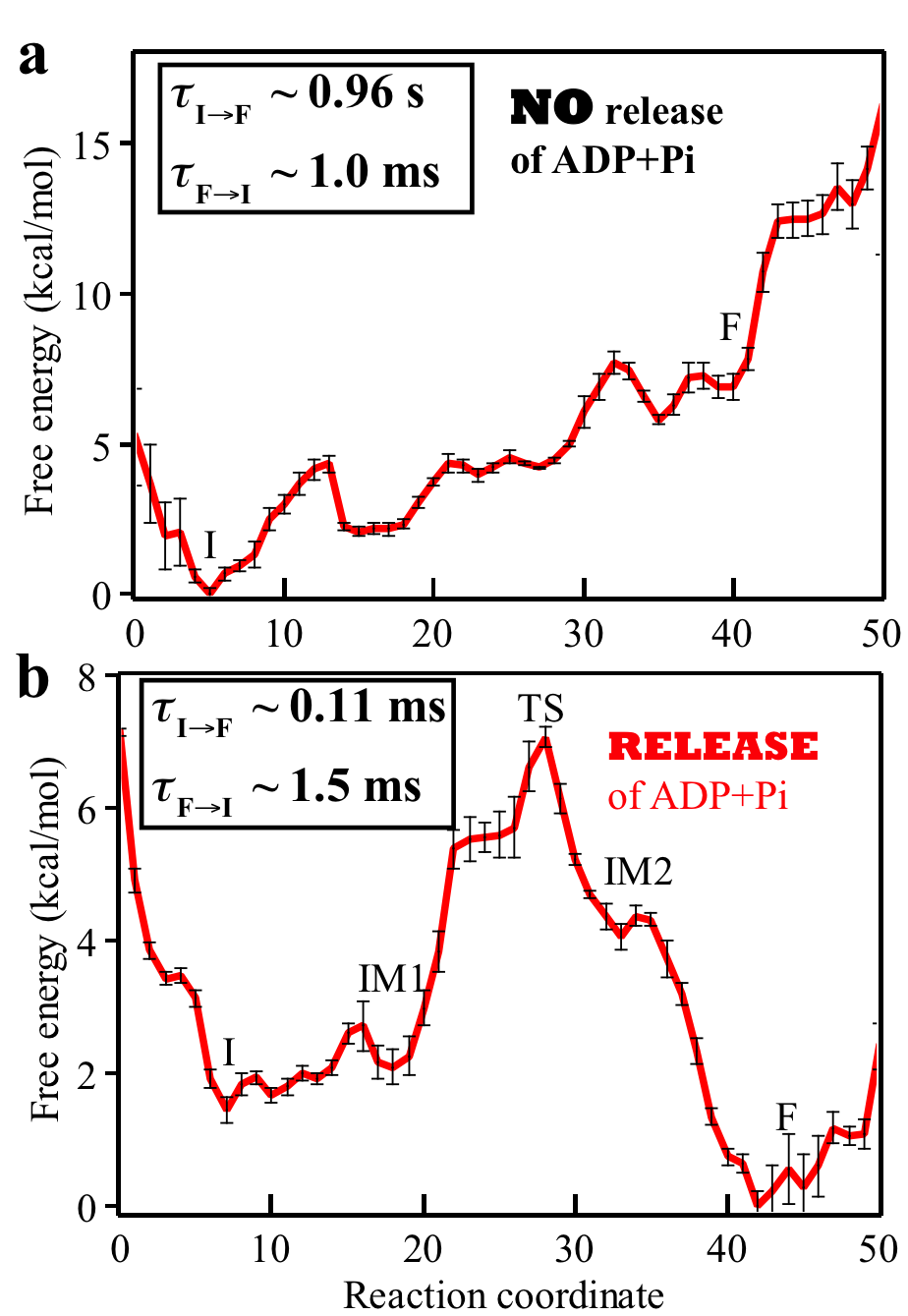
Fig. 4: Free energy profiles and mean first passage times characterizing the rotatory reaction step of Rho. The question addressed in the present study is how the free energy stored in the motor during the dwell phase is transferred to power substrate motion in the motor-action phase. Essentially we need to characterize conformational transitions of a motor in the motor-action phase. Due to the millisecond time-scale of the conformational transition along with a molecular motor's large size, straightforward MD simulations cannot reach the time scale of the transitions. To overcome this limitation, we carry out MD simulations employing the string method and the milestoning technique to characterize the conformational transition pathway of Rho's action phase along with associated free energy profile and kinetic rates in the Rho-RNA complex. For the methodological details of methods, see Supporting Information of [ Ma and Schulten, JACS, 2015 ]. Our implementation of the path sampling methods benefitted from a scalable multiple copy algorithm implemented in NAMD. The rotary reaction R → F (Fig. 3) is driven by ligand state changes of the ATP hydrolysis cycle. The state R represents a stable state corresponding to the crystal structure; in this state Rho does not transition spontaneously to F. As mentioned above, three subprocesses occurring during Rho's dwell phase are required to bring about a motor-action phase transition toward F (Fig. 2): (1) binding of ATP to the empty site, namely E → T; (2) release of "old" hydrolysis product (ADP + Pi), namely D → E; (3) formation of "new" hydrolysis product through the reaction ATP → ADP + Pi, namely T* → D*. D represents here a state of loosely bound ("old") product and D* a state of tightly bound ("new") product. The subprocesses (1, 2, 3) take place somewhere along the transition R → F. Based on experimental studies, subprocess (2) is considered rate limiting. Accordingly, we assume in our study that subprocesses (1, 3) take place at the very beginning of the R → F transition. To elucidate the rate-limiting nature of subprocess (2) we have studied the conformational transition pathways for Rho's motor-action phase for two cases: (a) hydrolysis product is not released from the m6/m1 interface binding pocket during the whole motor-action phase; (b) hydrolysis product (ADP+Pi) is released from the interface in the beginning of the motor-action phase. We have found the conformational transition pathways for Rho's motor-action phase in the two cases a and b (see the top and bottom plots in Fig. 4, respectively). The free energy and mean first passage times are presented in Fig. 4. A comparison of two cases shows clearly that the rotary reaction proceeds much better when the product release happens early on in the initial state I. Without product release the overall reaction is strongly endergonic, and the forward rate constant is 1000 times smaller than the reverse rate constant. After product release, the reaction becomes exergonic; the calculated mean first passage time (MFPT) from I to F is evaluated to be about 0.11 ms, consistent with the experimentally estimated value [ Adelman et. al, 2006, Mol. Cell ]. |
Relative motions of Rho subunits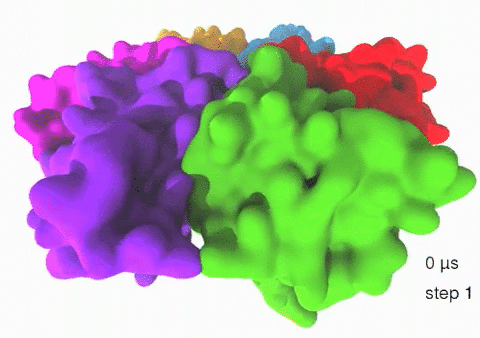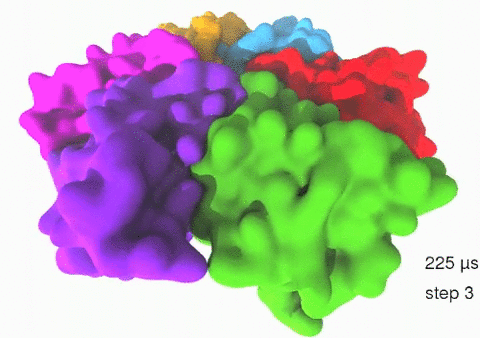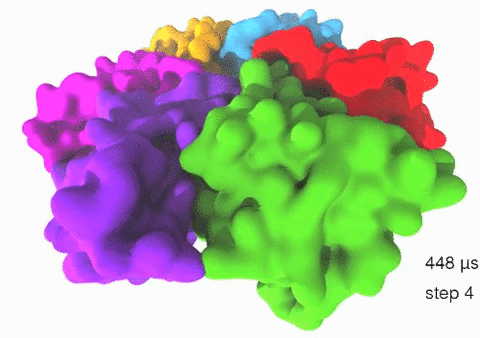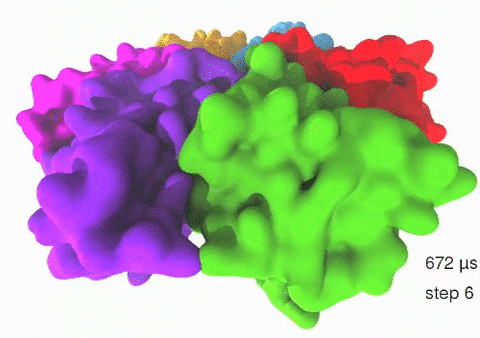 Movie 1: Relative motions of Rho subunits during six sequential rotary reaction steps.
(Download the movie here)
Movie 1: Relative motions of Rho subunits during six sequential rotary reaction steps.
(Download the movie here)
In the following we analyze the characteristics essential for Rho's overall motion along the transition pathway after product release. Movie 1 shows the overall translational motion of the six Rho subunits along the z-axis. In this movie, the scenario described above for one rotary reaction step (step 1 in Fig. 2, or equivalently R → F in Fig. 3) has been readily extended to an arbitrary number of such steps (e.g. steps 2, 3, ... in Fig. 2). The step 1 hexatuples of ligand states (T*T*TTED → DT*T*TTE) corresponding to the transition R → F can be continued by shifting the step 1 ligand states by one Rho subunit. For example, step 2 involves then ligand state changes DT*T*TTE → EDT*T*TT; ...; step 7 involves ligand state changes T*T*TTED → DT*T*TTE. Step 7 exhibits again the same ligand states as step 1; the shifts complete in step 6 a full cycle (360°) through all six Rho subunits. Thus Movie 1 shows the relative motion of six Rho subunits in a full cycle (six steps). For the sake of better visulization, we do not show the RNA here although all the simulations include the RNA. The first mean passage time (MFPT) in µs from the initial state to a state of a given frame is shown in the right-bottom corner; the MFPT values show the time evolution of the Rho-RNA conformational transitions. |
How unidirectional RNA translocation is coupled to Rho conformational transitionsThe coordinated motion of Rho's subunits serves the purpose of translocating the bound RNA along Rho's central pore. The translocation results from a coupling between Rho subunits and RNA, actually involving mainly interactions between six lysines K326 of subunits m1, m2, ..., m6 and the RNA backbone. Movie 2 highlights the important role for RNA translocation of the six K326 side chains accompanying the motion of the Rho subunits (in a full cycle as Movie 1). The motion of the lysines is well adjusted to the geometry of the bound RNA such that the RNA gets pulled in a linear direction through engagement with and disengagement from the backbone phosphate groups; the RNA adopting a helical geometry in its bound state does not rotate as it is translocated relative to Rho. The side chains of the lysines K326 of subunits m1, m2, m3 and m4 are shown in different opaque colors (same as those in Movie 1). The RNA is shown in pink cartoon representation. More details of the transition pathway and free energy landscape for subunits-RNA interactions can be found here. |
Allosteric pathway that couples RNA translocation to hydrolysis product release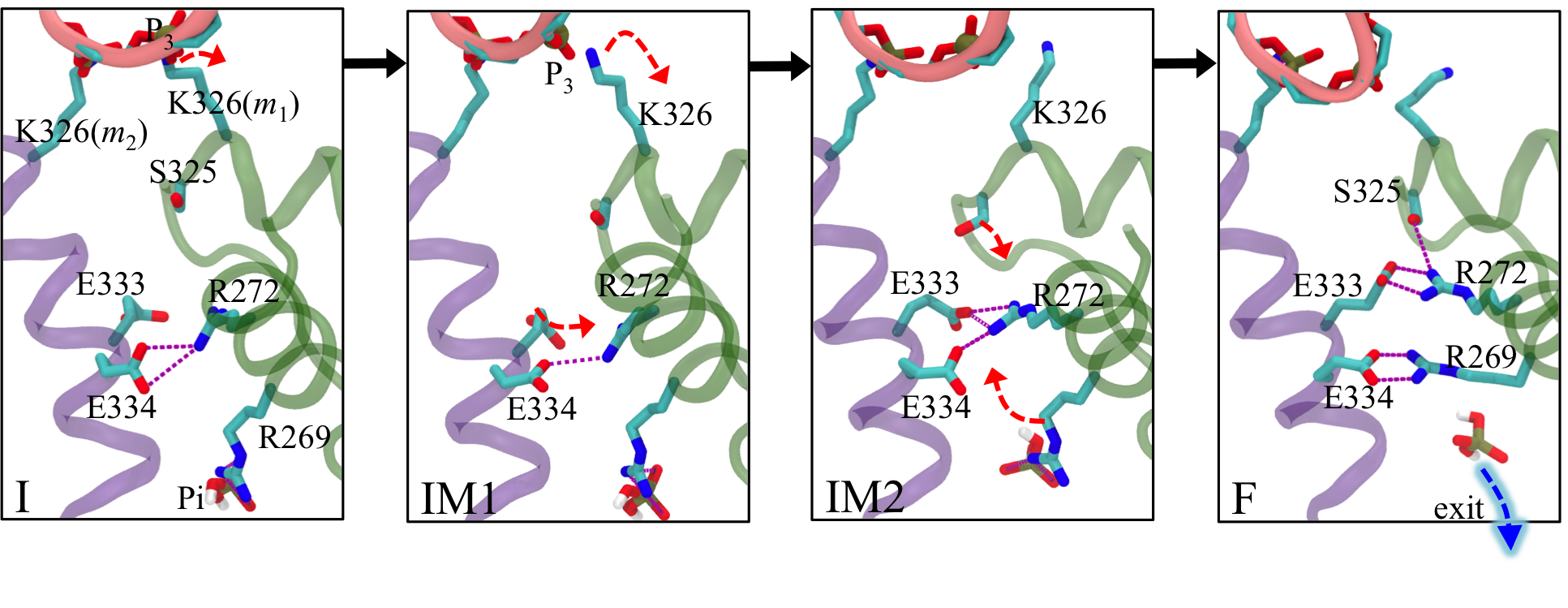 Fig. 5: Allosteric coupling between RNA translocation and Pi of the hydrolysis product. Transitions I → IM1 → IM2 → F are shown as black arrows.
Fig. 5: Allosteric coupling between RNA translocation and Pi of the hydrolysis product. Transitions I → IM1 → IM2 → F are shown as black arrows.
RNA translocation after step 1 requires at the beginning of step 2 the release of hydrolysis product ADP + Pi at the m1/m2 interface. In fact, when we assumed for step 1 that the respective release of "old" product at the m6/m1 interface does not take place, the rotatory reaction step is endergonic and cannot proceed (see case-a in Fig. 4). Likewise, for step 2, the hydrolysis product at the m1/m2 interface must be released before the next step of RNA translocation can proceed. For this purpose the hydrolysis product at the m1/m2 interface, which we termed "new" product, must experience a change from being tightly bound (D* state) to being loosely bound (D state) and, therefore, ready for subsequent release. One may hypothesize then that the initiation of step 2 in the form of a D → E transition at the m1/m2 interface during the next dwell phase should actually be prepared when state F is reached in step 1. It appears that such lockstep synchronization is indeed achieved by Rho through an allosteric pathway that links disengagement of K326(m1) from RNA backbone phosphate P3 to a liberation of inorganic phosphate Pi from a salt bridge with R269(m1) (see Fig. 5). This liberation loosens the binding of Pi.
PublicationsMechanism of Substrate Translocation by a Ring-Shaped ATPase Motor at Millisecond Resolution. Wen Ma and Klaus Schulten, Journal of the American Chemical Society, 137 (8), 3031-3040, 2015. Supporting Information: Mechanism of Substrate Translocation by a Ring-Shaped ATPase Motor at Millisecond Resolution. Wen Ma and Klaus Schulten, Journal of the American Chemical Society, 137 (8), 3031-3040, 2015.
InvestigatorsPage created and maintained by Wen Ma. |

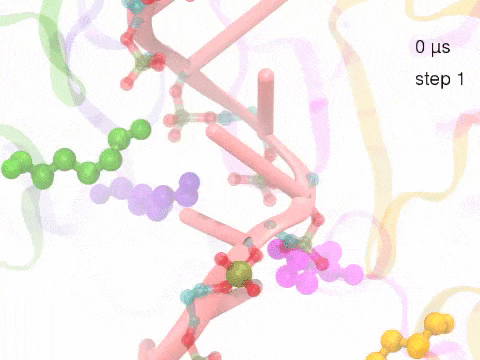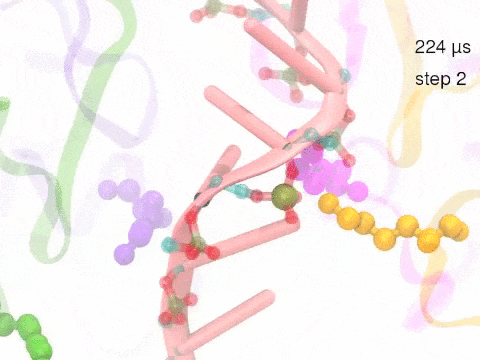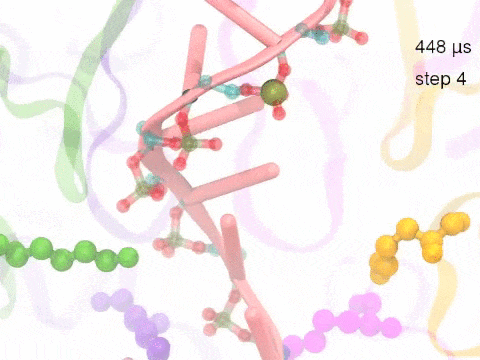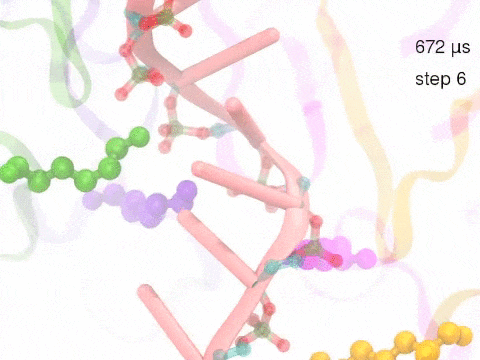 Movie 2: Six lysines from Rho subunits collaboratively translocate RNA. (Download the movie
Movie 2: Six lysines from Rho subunits collaboratively translocate RNA. (Download the movie 

