Bringing Physics to Life
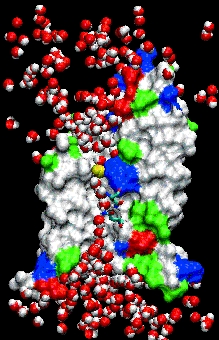 Gatekeeper. Aquaporin allows water molecules (red and
white) but not protons to pass through its central pore (see
movie).
Gatekeeper. Aquaporin allows water molecules (red and
white) but not protons to pass through its central pore (see
movie).
In 1944, the physicist Erwin Schrödinger, the founder of modern quantum mechanics, published a short book that changed the course of modern biology.
"What is Life?" he asked famously in his title. Could the events inside a living organism be explained solely by physics and chemistry? Yes, they could, Schrödinger answered. "The obvious inability of present-day physics and chemistry to account for such events," he wrote, "is no reason at all for doubting that they can be accounted for by those sciences."
It's a sentiment that has lured generations of physical scientists to biology.

For the past half-century, researchers have applied the rigorous tools of physics to help answer Schrödinger’s question and uncover the fundamental mechanisms of life. A series of Nobel prizes attests to their success: Francis Crick, James Watson and double helical DNA; Max Perutz and myoglobin; Hartmut Michel and photosynthetic reaction centers, Peter Agre and water channels - the list goes on. But some of the most exciting challenges remain, says Klaus Schulten, professor of physics and director of the Beckman Institute’s Theoretical and Computational Biophysics Group (TCBG).
Since 1989, when Schulten founded the group, TCBG researchers have been meeting those challenges. They’ve probed how cells harvest energy from sunlight, regulate genes, control water flow into and out of cells, pack DNA into chromosomes, remove excess cholesterol from the bloodstream, and more.
In so doing, says Schulten, they established themselves as biophysicists, and "altered the course of their lives."
Teamwork and Synergy
Schulten's vision and enthusiasm guide TCBG research. From the beginning of his career, he says, he was interested in the physical mechanisms by which cellular processes work. "I wanted to understand them rigorously," he says.
TCBG researchers achieve that rigor by combining theoretical and modeling work, and by collaborating with experimental biologists in the University of Illinois at Urbana-Champaign physics, chemistry, biochemistry, and cell and molecular biology departments, and at other universities and laboratories worldwide. Most TCBG researchers also consult with the five professional programmers in the group. "If there's any [problem] you go three doors down. It can't get any better in terms of getting work done," says TCBG graduate student Barry Isralewitz.
Even the building was designed with collaboration in mind. The TCB group is housed in a wing of the Beckman Institute, an attractive six-story limestone-and-brick building set on a broad green quadrangle. Researchers gather in conference rooms, nosh in the first-floor café in the spacious atrium, or gather to brainstorm in small lounge areas on the walkways that cross above it.
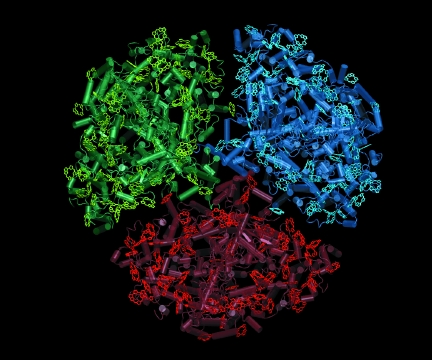 Tuning in. Trimeric photosystem I from the cyanobacterium
Synechococccus elongatus uses chlorophyll networks, to harvest energy from
sunlight.
Tuning in. Trimeric photosystem I from the cyanobacterium
Synechococccus elongatus uses chlorophyll networks, to harvest energy from
sunlight.
Despite the collaborative approach, each researcher creates his or her own niche, rather than being a cog in a big machine, says Elizabeth Villa, a TCBG graduate student in U of I's Center for Biophysics and Computational Biology. "You get to know people, you collaborate with people, but it’s your work," she says.
Schulten's approach and the state-of-the-art facilities have helped the TCB group grow into one of the world's top biophysics laboratories, with more than $2 million annually in external funding. The group operates the National Institutes of Health Resource for Macromolecular Modeling and Bioinformatics, a unique center that develops new algorithms and computing tools for structural biology. The group includes four other U of I faculty - computer science professors Robert Skeel and Laxmikant Kale, chemistry professors Todd Martinez and Zan Luthey-Schulten—as well as Emad Tajkhorshid, assistant director of the NIH center. More than a dozen of Schulten’s students have landed faculty positions at top institutions, including University of Illinois, Chicago; University of California, Irvine; and Stanford University.
Molecules in Motion
The TCB group is well known for its successful simulations of macromolecules at work. Structural biologists have long determined the detailed three-dimensional structures of proteins and the cell's other macromolecules, pinpointing the position of the thousands of component atoms. Those structures offer clues about how proteins act as motors, channels, solar cells, or genetic switches. "But the structure is just a starting point," Schulten says. To begin explaining how a protein functions, he explains, it's important to see it move.
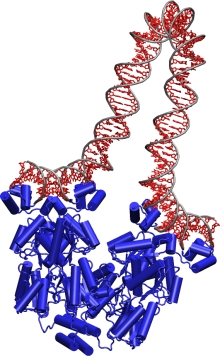 Getting a grip. TCBG researchers probe how the lac repressor (blue)
grasps DNA and bends it like a pretzel.
Getting a grip. TCBG researchers probe how the lac repressor (blue)
grasps DNA and bends it like a pretzel.
To do that, TCBG researchers have created software that simulates macromolecules at work. For example, the molecular graphics program Visual Molecular Dynamics (VMD), which allows researchers to create three-dimensional movies of macromolecules, has more than 38,000 registered users. The group’s popular molecular-dynamics program NAMD enables simulations using large-scale parallel machines with thousands of processors. Group members have routine access to some of the world’s largest computers at the nearby National Center for Supercomputer Applications, which is one of just three National Science Foundation-funded supercomputing centers in the country.
The computational firepower and know-how gives the TCB group a competitive edge, Tajkhorshid says. TCBG researchers can simulate many nanoseconds of motion of huge membrane proteins containing more than 300,000 atoms - routinely shedding new light on how large proteins work.
To illustrate, Tajkhorshid hands a visitor a set of stereoglasses, taps some keys on a computer, and projects a movie onto a huge screen. The screen looks like the darkened window of an aquarium; the strange off-white creature inside is a scale model of a channel protein called aquaporin that controls the flow of water into and out of cells.
He taps a few keys, and dozens of rounded red and white water molecules swarm around the much-larger aquaporin channel like bees; a line of them squeeze single-file through a narrow hole in the channel’s center. Halfway through, each water molecule flips 180 degrees.
The aquaporin simulation, which Tajkhorshid, Schulten, and their experimental colleagues published in Science in 2002, was "the first complete representation of transport of material across the membrane in full atomic detail," Tajkhorshid says. [See Structure, Dyanmics, and Function of Aquaporins research page here]
It also solved a longstanding mystery about aquaporins that the detailed atomic structure had not. Scientists had wondered how aquaporins allowed water molecules but not protons to flow through, even though protons are much smaller. As it turned out, flipping the water molecules around prevented protons from traveling through the channel on what amounts to a wire made of water. That, in turn, helps the microbe keep its membrane charged up like a solar cell, which provides the fuel it needs to stay alive.

Physics at Work
When Schrödinger first speculated on the nature of life, there was a wide disconnect between physics and biology. No one knew the physical nature of a gene, the term molecular biology had yet to be invented, and many of the smartest scientists went into elementary particle physics. No more, says Schulten.
Today, physical life sciences is a mature field and a competitive one, offering opportunities to physicists of all stripes. In the TCB group alone, researchers apply quantum mechanics, quantum electronics, condensed matter physics, optics, statistical mechanics, and classical mechanics. "You can do exciting physics in life sciences," Schulten says, "but in systems that are eminently relevant" to mankind.
Melih Sener, a TCB postdoctoral researcher from Turkey, does just that.
Recently he applied a modeling method he'd learned as a graduate student in theoretical high-energy physics - random-matrix theory - to shed light on how a photosynthetic bacterium harvests energy from sunlight. The bacterium, called Synechococcus elongatus, contains photosynthetic machinery like that of higher plants, including a large cloverleaf-shaped protein complex with three copies of a membrane protein called photosystem I. Each copy of photosystem I contains 96 chlorophyll molecules, which act together as an antenna to trap light energy and use it to "charge the [cell] membrane up like a solar cell," Sener says.
Other researchers had determined the three-dimensional structure of the protein, pinpointing the position and orientation in space of each of its chlorophylls, which enabled Sener, Schulten, and their coworkers to calculate how the chlorophylls would harvest light energy when the chlorophylls were frozen in place. But life happens at room temperature, where the 96 chlorophylls are constantly jostling, twisting, and turning.
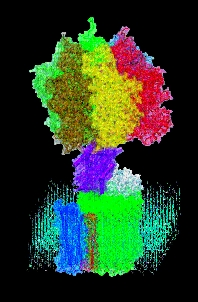 Proton power The F0-F1 ATP synthase harvests
energy from a transmembrane proton gradient, using it to make ATP.
Proton power The F0-F1 ATP synthase harvests
energy from a transmembrane proton gradient, using it to make ATP.
The researchers used random-matrix theory to calculate how that motion affects the way ensembles of chlorophyll trap light energy. That, in turn, helped reveal that nature had arranged the 96 chlorophylls for optimal activity, and that the jostling has little effect on how well the antennae harvests light energy, according to results published in 2002 in the Journal of Physical Chemistry B.” In some cases we can bring the heavy artillery from theoretical physics to bear upon biological problems,” Sener says.
Other branches of physics also come in handy. Isralewitz, fellow TCBG graduate student Markus Dittrich, and postdoctoral researcher Alek Aksimentiev apply the tools of computer simulation, statistical mechanics and quantum mechanics to learn how cells tap stored solar energy to drive a molecular turbine called F1-F0 ATP synthase that makes ATP. And TCBG graduate student Villa uses a cutting-edge modeling method called multiscale modeling to determine how a protein called the lac repressor grabs onto DNA and bends it like a garden hose, preventing it from expressing its genes.
Ongoing work in the TCB group focuses on a new frontier of physical life sciences - how individual components of the cell work together. Engineering is another frontier. By understanding the design principles of cellular machinery, Schulten says, "we are learning from nature how to design new devices" that electrically record single molecules and perform other tasks. For engineers and physicists, he concludes, "much of the future lies in life sciences."
It's all a long way from Schrödinger. But scientists are still pressing forward to bring the best of physics to biology. What is life? Six decades later, the question remains compelling.
Article: Dan Ferber
Design: Amy Harten
Photos: John Dessen
Web Design: Tim Skirvin
Beckman Institute for Advanced Science & Technology
University of Illinois at Urbana-Champaign
405 N. Mathews Ave.
Urbana, IL 61801
http://www.ks.uiuc.edu/



